Complete Overview of Small RNA Sequencing: Methods, Workflow, Platform, and Applications
Small RNA sequencing has emerged as a potent instrument within the realm of molecular biology, affording a nuanced exploration into the domain of non-coding RNAs (ncRNAs) and their multifaceted roles. Through its capacity to discern and enumerate minute RNA species, including but not limited to microRNAs (miRNAs), small interfering RNAs (siRNAs), and transfer RNA-derived small RNAs (tsRNAs), small RNA sequencing has engendered a paradigm shift in our comprehension of gene modulation, cellular dynamics, and pathological pathways.
Introduction
Small RNAs, typically spanning from 18 to 200 nucleotides, wield profound influence over gene expression regulation, RNA interference, and epigenetic modifications. Within the vast landscape of small RNAs, miRNAs and siRNAs have seized considerable interest for their pivotal roles in post-transcriptional gene silencing and mRNA degradation. Furthermore, tsRNAs, arising from transfer RNAs, have recently surfaced as novel regulatory entities entangled in an array of physiological and pathological processes.
Small RNA sequencing, commonly referred to as sRNA-seq, facilitates the thorough examination of small RNA populations present within biological specimens. Through the utilization of high-throughput sequencing methodologies, investigators gain access to the intricate milieu of small RNAs, allowing for the elucidation of their expression profiles, sequence diversities, and functional ramifications. This manuscript offers a comprehensive exposition on the methodologies, workflows, sequencing platforms, and myriad applications pertinent to small RNA sequencing across various research realms.
Why Small RNA Sequencing?
sRNA sequencing possesses a multitude of advantages over classical approaches such as northern blotting and quantitative PCR, which serve as staples in small RNA analysis. A primary benefit residing in sRNA-seq is the capacity to garner a comprehensive purview of global small RNA populations. This enables detecting elusive or scarcely found species - entities that might otherwise evade discovery via conventional methodologies.
In addition, sRNA sequencing facilitates the recognition and identification of new, undescribed small RNAs and their isoforms, thus offering a springboard for unearthing hitherto undetected regulatory constituents.
sRNA-seq additionally brings forward a treasury of quantitative insights regarding small RNA expression strata, offering an impeccable precision in quantifying transcript loads across an array of contrasting experimental conditions or biological specimens. This wealth of quantitative information can play an instrumental role when engaged in studies warranting a deep examination of dynamic alterations in small RNA expression, in scenarios that may include development processes, the journey of disease progression, or evaluating responses to therapeutic interventions.
Moreover, sRNA sequencing lays the groundwork for characterizing the convoluted pathways and regulatory mechanisms connected to small RNA biogenesis. By aligning small RNA sequences to their genomic origins and antecedent molecules, scholars can disentangle the complexities tied with small RNA handling, refining, and turnover. This accords pivotal mechanistic insights into the expansive networks of gene regulation, underscoring the importance of sRNA sequencing in contemporary biological research.
Small RNA Sequencing Methods
Small RNA sequencing methodologies comprise an array of techniques tailored for the analysis of small RNA molecules. These methods exhibit diversity in their approaches to small RNA library preparation, adaptor ligation, size selection, reverse transcription, and amplification. The selection of a particular method hinges upon the precise research objectives, the nature of small RNAs under scrutiny, as well as the resources and expertise at hand.
Some of the commonly used methods for small RNA sequencing include:
Polyadenylation-based methods
Polyadenylation-based methods constitute a vital protocol in the analysis of sRNAs, including but not exclusive to miRNAs. This approach fosters an efficacious isolation of specific sRNA classes due to a fundamental, enzymatic polyadenylation process.
The procedure commences with the biochemically induced modification of sRNAs through the attachment of poly(A) tails at their 3' extremities. This significant adaptation creates a uniform recognition domain for the subsequent ligation of adaptors.
Upon polyadenylation, adaptors, embedded with oligo-dT sequences, are fastened to the polyadenylated sRNAs. These adaptors not only function as priming sequences for complementary DNA synthesis but also ensure the selective gathering of polyadenylated sRNAs. The propensity of the oligo-dT sequences in the adaptors to specifically bind to the poly(A) tails promotes the preferential capture of miRNA and other similar homologs.
The pros of polyadenylation-based protocols lay in their propensity to selectively augment miRNAs - most of which bear poly(A) tails. The selective enhancement facilitates heightened detection sensitivity during sRNA sequencing. Furthermore, deploying polyadenylation-based methods can restrict the identification of alternative RNA types void of poly(A) tails. This selective parsing downplays the sampling noise, thus, improvising the specificity within the realm of miRNA detection.
Direct Ligation-Based Methods
Complimenting this approach are Direct Ligation-Based Methods, an alternative mode of operation in library preparation for sRNA studies. This method is distinguished by its elimination of the polyadenylation process, a ubiquitous step in many other strategies.
The conveniences offered by this method stem from the immediate ligation of adaptors to sRNA ends, thus excluding any requirement for preceding polyadenylation. From this primary distinction derives a host of benefits for scientific inquiry into varied sRNA types, including miRNAs, piRNAs, and tRNAs.
Notably, direct ligation methods, by omitting the polyadenylation step, respect the original properties of the many species of sRNA and facilitate a broad and comprehensive study of the multiple RNA populations within a given sample.
Direct ligation of adaptors to sRNA has several advantages in experimental execution and potential results. It streamlines the preparation process by removing the need for successive enzymatic actions. This reduction in steps considerably minimises the opportunities for bias and artificial influence, which might otherwise be introduced during the polyadenylation process. Moreover, this strategy ensures the proficient capture and subsequent detection of sRNAs sans polyadenylation attribute, such as piRNAs and tRNAs. These particular sRNAs are well-known for their key roles across diverse cellular processes.
As another noteworthy advantage, direct ligation-based methods provide versatility in study design and subsequent analysis, enabling researchers to attune protocol development to specific research questions and potentially varied sample types. Given its versatile nature, these strategies significantly enrich research utility, making them highly suitable for investigating intricate sRNA populations across diverse biological conditions, be they developmental, diseased states, or in response to environmental stimulus.
Size selection methods
The utilisation of size selection strategies proves indispensable in the realm of sRNA research, primarily facilitating the enrichment of distinct sRNA subsets within intricate RNA mixtures. Size selection methods expedite the segregation of the research-interest sRNAs, generally ranging 18 to 30 nucleotides, from the composite pool of whole RNA.
Amidst a spectrum of available methodologies, gel electrophoresis emerges as a predominantly deployed technique for size selection in sRNA investigations. In electrophoresis, all-encompassing RNA specimens are introduced into a denaturing polyacrylamide gel and electrophoresed under conditions instigating RNA denaturation. Consequently, based on their respective sizes, RNA molecules progress through the gel matrix, with a rapid migration rate for smaller entities as compared to the larger ones. The traversal of RNA molecules through the gel results in the banding patterns which correspond to particular size domains, inclusive of the sought-after sRNA fraction. These bands are detectable using nucleic acid stains or florescent dyes, then extracted from the gel, ensuing the elution and retrieval of sRNAs for downstream methodologies, such as library preparation for next-generation sequencing.
Parallelly, size-exclusion chromatography presents a high-throughput and scalable methodology for sRNA size selection. Using this methodology, comprehensive RNA specimens are introduced into a size-exclusion chromatography column packed with porous resin beads. As the sample navigates through the column, molecules segregate according to their size. Smaller entities tread more convoluted paths through the matrix resin, rendering slower elution times. Researchers can thus effectively isolate and concentrate the sRNA population of interest by gathering fractions corresponding to the desired size domain, such as the 18-30 nucleotide window for sRNAs.
Both gel electrophoresis and size-exclusion chromatography afford scientific investigators the latitude to customise size selection parameters anchored on precise experimental stipulations. For example, practices in gel percentage selection, execution conditions, and staining methodologies can be perfected to attain exact separation and visualisation of sRNAs. Similarly, the choice of column matrices and elution buffers in size-exclusion chromatography can be tailored to augment efficiency and purity of sRNA segregation.
Modified methods for specific small RNA classes:
Conventional strategies frequently integrate specific capture probes or primers during the library preparation phase, which are engineered to selectively hybridize with target small RNA sequences. This effectively enhances targeted enrichment from the general RNA cohort. This targeted approach not only refines the complexity of the sample but also improves detection sensitivity, thereby enabling scientific focus on a particular subtype of RNA during sequencing.
In addition, modified methodologies may utilize adaptors or linkers that have been optimized according to the unique structural characteristics of the specific small RNAs under investigation. In certain instances, these specially-designed adapters can vastly improve the ligation or amplification efficiency of miRNAs or PIWI-interacting RNAs (piRNAs). Concurrently, the introduction of blocking oligonucleotides can effectively inhibit any unwanted non-target RNA interactions, providing further refinement to the method's specificity.
Modified approaches may also feature the incorporation of size-selection or fractionation steps aimed at isolating a targeted sRNA class from the larger, diverse pool of RNA. This is extremely beneficial when it comes to enriching miRNAs or piRNAs, which are typically confined to specific size ranges. Techniques such as gel electrophoresis or size-exclusion chromatography can be employed to separate targeted small RNA factions, thereby facilitating a more streamlined and efficient sequencing process.
Service you may intersted in
Small RNA Sequencing Steps
Sample QC and RNA Integrity Assessment
Before embarking on small RNA sequencing, it is essential to assess the quality and integrity of RNA samples to ensure optimal sequencing outcomes. Quality control (QC) metrics, such as RNA concentration, purity, and integrity, are evaluated using spectrophotometry, fluorometry, and gel electrophoresis. High-quality RNA samples with intact small RNA populations are essential for accurate small RNA sequencing.
Library Preparation
The role of library preparation in small RNA sequencing is absolutely critical; the quality and accuracy of the consequent sequencing data hinge on this. Effective library preparation procedures ought to ensure an efficient capture of small RNA molecules, minimize any potential biases and enable multiplexing for elevated throughput.
The process of Small RNA-seq commences with the isolation of small RNA molecules from total RNA strands or enriched RNA fractions. The isolated small RNAs then undergo conversion into a sequencing library via several steps. These stages include: adapter ligation, reverse transcription and PCR amplification. A range of commercial kits are accessible for small RNA library preparation, each presenting their own specific advantages and potential biases.
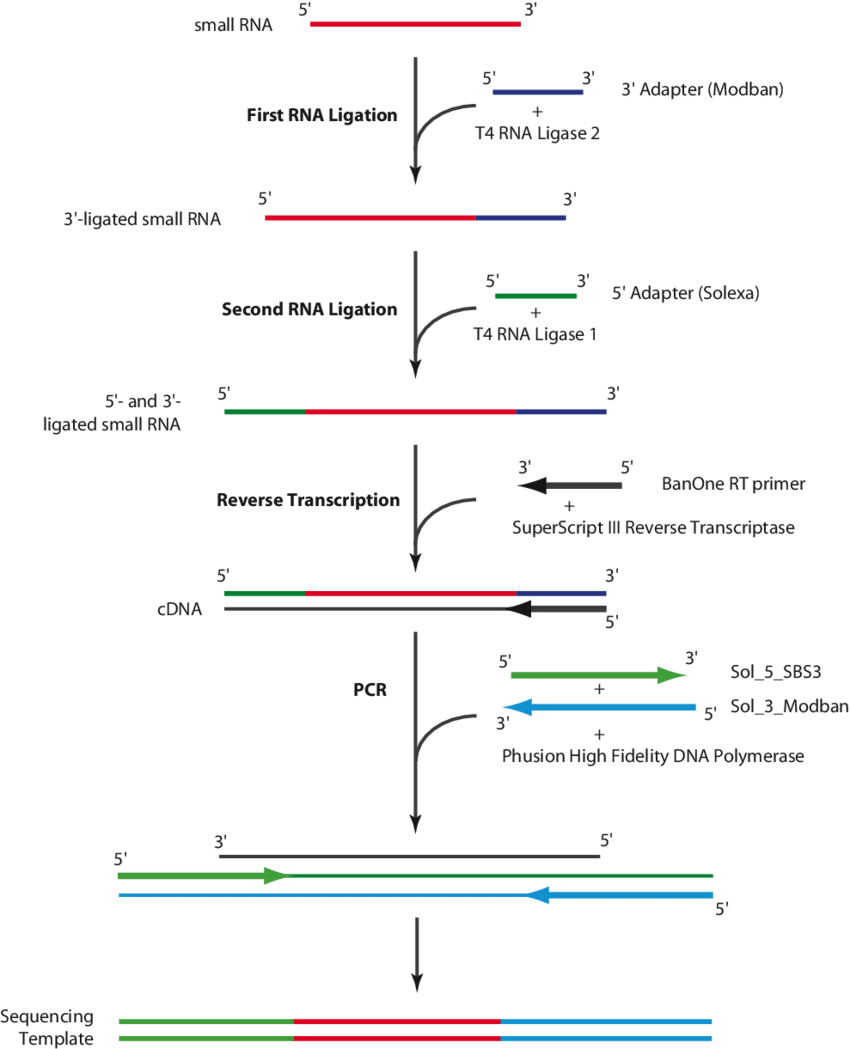 Small RNA library preparation (Valérie Gausson et al,. 2011)
Small RNA library preparation (Valérie Gausson et al,. 2011)
Size Selection
Following adapter ligation, size selection is often performed to enrich for small RNA fragments of the desired length range. Gel electrophoresis or bead-based purification methods are commonly used for size selection, allowing researchers to remove unligated adapters and other contaminants from the library.
Reverse Transcription and PCR Amplification
After size selection, the small RNA fragments with ligated adapters are reverse transcribed into cDNA, which serves as the template for PCR amplification. PCR amplification enriches the small RNA-seq library and introduces index sequences or barcodes for sample multiplexing.
Sequencing
The prepared small RNA-seq libraries are then subjected to high-throughput sequencing using next-generation sequencing platforms, such as Illumina or Ion Torrent. During sequencing, the adapters serve as priming sites for the sequencing reaction, allowing the determination of the small RNA sequences and their abundance.
Data Analysis
Following sequencing, the generated raw data undergoes a series of bioinformatics analyses to preprocess, map, quantify, and annotate small RNA sequences. Bioinformatics pipelines for Small RNA sequencing data analysis typically include quality control, adapter trimming, read mapping to reference genomes or small RNA databases, and expression quantification.
Various software tools and algorithms are available for small RNA data analysis, including Bowtie, BWA, STAR, miRDeep2, and DESeq2. These tools enable researchers to identify known miRNAs, predict novel miRNAs, quantify expression levels, detect differential expression, and annotate small RNA sequences based on genomic features.
Additionally, advanced bioinformatics approaches, such as small RNA pathway analysis, target prediction, and functional enrichment analysis, can provide deeper insights into the biological functions and regulatory roles of small RNAs. Integration of small RNA sequencing data with other omics data, such as mRNA-seq and ChIP-seq, further enhances our understanding of gene regulatory networks and signaling pathways.
To learn more, please refer to "The Challenge and Workflow of Small RNA Sequencing"
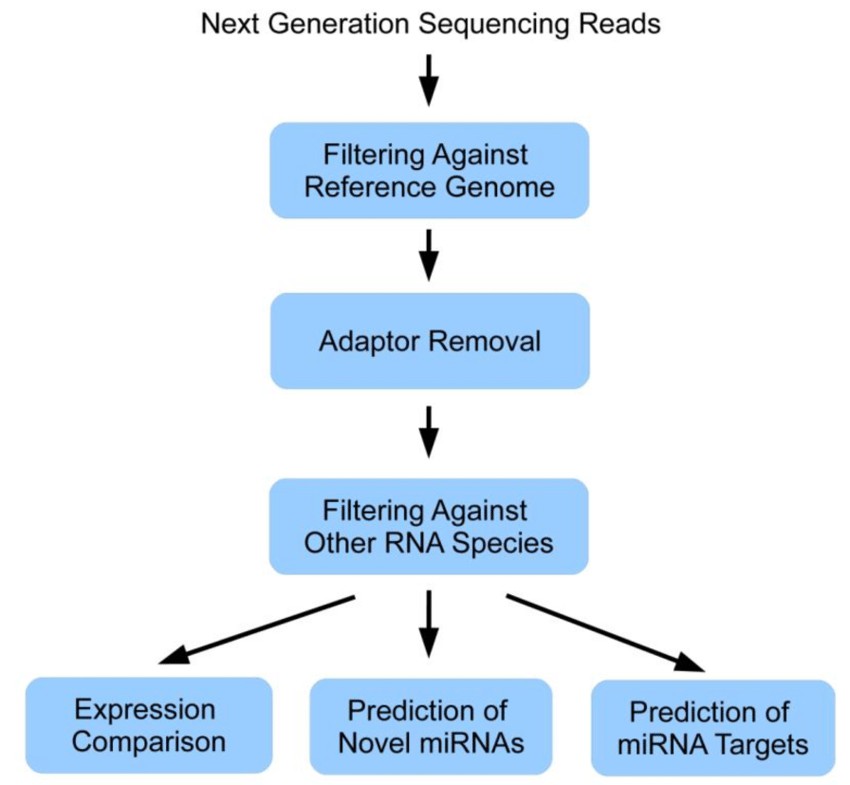 Data analysis workflow of a miRNA next generation sequencing experiment. (Susanne Motameny et al,. 2010)
Data analysis workflow of a miRNA next generation sequencing experiment. (Susanne Motameny et al,. 2010)
Workflow of Data Analysis
| Workflow of Data Analysis |
| Preprocessing and Quality Control |
| The initial stage of small RNA sequencing data analysis involves preprocessing and quality control. This includes adapter trimming, read filtering, and elimination of low-quality reads to uphold the integrity of sequencing data. CD Genomics delivers comprehensive bioinformatics services for preprocessing and quality control, ensuring that sequencing data adheres to the highest standards of precision and dependability. |
| Read Mapping and Quantification |
| After preprocessing, the subsequent phase encompasses read mapping and quantification, during which sequencing reads are aligned to a reference genome or transcriptome to pinpoint the origin of small RNA molecules. CD Genomics employs cutting-edge alignment algorithms and quantification techniques to precisely map reads and quantify small RNA expression levels across diverse samples and conditions. |
| Advanced Data Analysis Techniques |
| Beyond basic read mapping and quantification, advanced data analysis methods are utilized to glean deeper insights from small RNA sequencing data. |
| Differential Expression Analysis |
| Differential expression analysis scrutinizes the expression levels of small RNA molecules across different experimental conditions, such as healthy versus diseased samples or treated versus untreated cells. By pinpointing differentially expressed small RNAs, researchers can unearth potential biomarkers or regulatory pathways associated with specific biological processes or diseases. |
| Small RNA Isoform Analysis |
| Small RNA isoform analysis delves into characterizing the sequence diversity and isoform variants within small RNA populations. CD Genomics provides specialized tools for identifying and quantifying small RNA isoforms, empowering researchers to explore the functional diversity and regulatory intricacies of small RNAs in greater depth. |
To learn more about Small RNA Data Analysis, please click "Bioinformatics Analysis of Small RNA Sequencing"
Small RNA Sequencing Platforms
There are a number of Next-Generation Sequencing platforms routinely leveraged in executing Small RNA sequencing, notable among them: Illumina, and Ion Torrent. Each individual platform possesses inherent advantages over the others in several respects, encompassing parameters such as read length, sequencing depth, throughput capability, and cost efficiency.
Predicated on reversible terminator chemistry, Illumina sequencing represents the most prevalent platform in the field of small RNA sequencing. Its established prominence can be attributed to its distinctive combination of high sequencing accuracy, scalability, and cost-effectiveness. By utilizing Illumina sequencing, researchers have the ability to produce millions of short reads, ranging typically from 50 to150 nucleotides in length, within a single sequencing run. Consequently, this facilitates an extensive coverage of small RNA populations.
Contrarily, the Ion Torrent sequencing platform harnesses a semiconductor-based method for detecting hydrogen ions released during the process of DNA synthesis. Despite boasting expedited turnaround times and simplified workflows, the Ion Torrent approach is more susceptible to homopolymer errors. Moreover, it could potentially deliver a lower sequencing accuracy when juxtaposed with Illumina sequencing.
Small RNA Sequencing on Illumina Platforms
Illumina sequencing has revolutionized the field of genomics with its high-throughput, accuracy, and cost-effectiveness. When applied to Small RNA sequencing, Illumina platforms offer several advantages, making them the preferred choice for many researchers.
Advantages of Illumina Sequencing for Small RNA:
| High Throughput | Illumina sequencers, such as the MiniSeq System, enable the simultaneous sequencing of thousands to millions of small RNA molecules in a single run. This high throughput allows for comprehensive profiling of small RNA populations from diverse biological samples. |
| Accuracy | Illumina sequencing delivers highly accurate base calls, ensuring reliable detection of small RNA sequences and their associated modifications. This accuracy is essential for identifying rare or low-abundance small RNA species and for quantifying expression levels with precision. |
| Short-Read Sequencing | Illumina platforms generate short-read sequences, typically ranging from 50 to 300 nucleotides in length. While this may limit the detection of longer RNA isoforms or structural variants, it provides excellent coverage of small RNA populations and facilitates mapping to reference genomes or small RNA databases. |
| Cost-Effectiveness | The per-base sequencing cost of Illumina platforms is lower compared to other sequencing technologies, making it a cost-effective solution for small RNA sequencing studies, especially when analyzing large sample cohorts or conducting high-depth sequencing experiments. |
| Established Protocols | Illumina offers well-established library preparation protocols and analysis pipelines specifically designed for small RNA sequencing. These standardized workflows streamline the experimental process and ensure reproducible results across different research projects. |
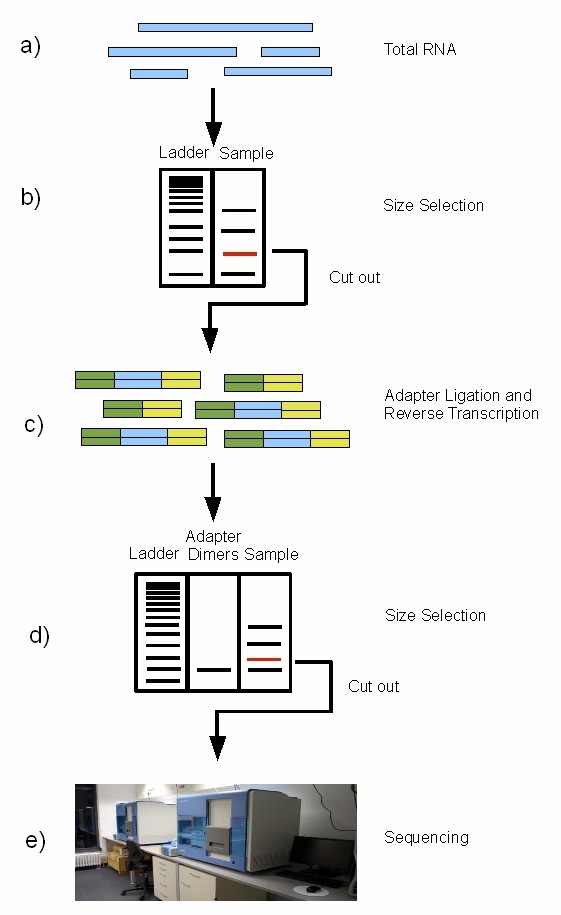 miRNA sequencing procedure on the Illumina (Susanne Motameny et al,. 2010)
miRNA sequencing procedure on the Illumina (Susanne Motameny et al,. 2010)
Applications of Small RNA Sequencing in Biology
Small RNA sequencing represents a revolutionary technological advancement with pervasive influence traversing a multitude of scientific disciplines. The array of its applications accentuates its crucial position in propelling the frontiers of biomedical research, environmental science, agricultural biotechnology, and personalized medicine. Its ability to act as a catalyst for innovation and discovery is clearly displayed in the relentless pursuit of enhanced human health and the sustainability of our environment. This cutting-edge sequencing method stands as a testament to the strides made in integrating complex scientific research with practical applications, signifying a tangible shift towards more effective scientific investigation and implementation.
1. Biomarker Discovery:
Serving as the backbone of biomarker discovery, small RNA sequencing assists in uncovering the roles of miRNAs, piRNAs, and other small RNA classes implicated in a wide array of diseases. This is achieved through the contrastive analysis of small RNA expression profiles within healthy and pathogenic tissues. Researchers, thereby, can pinpoint potential biomarkers vital for early disease detection, prognosis, and therapeutic stratification.
2. Disease Research and Pathway Analysis:
The use of small RNA sequencing is revolutionizing our knowledge base pertaining to the etiological underpinnings of a multitude of diseases. It enables researchers to uncover differential patterns of small RNA expression, thereby illuminating intricate regulatory networks divergent in health and disease. The critical pathophysiological pathways identified through this approach not only offer a comprehensive understanding of disease onset and progression, but also pave the avenue for the discovery of novel efficacious therapeutic targets. Thus, the application of small RNA sequencing is a crucial arbiter for the advancement of precision medicine, fostering a more profound comprehension of disease pathogenesis and its subsequent application in tailored therapeutic strategies.
3. Functional Genomics:
The technology of Small RNA sequencing plays a central role in illuminating the functional importance of small RNAs in the regulation of genes and intracellular operations. By conducting comprehensive characterizations of small RNA populations, scientists are enabled to pinpoint key regulatory entities, delve into the study of post-transcript gene regulation, and investigate the intricacies of RNA-mediated mechanisms involved in cellular development, differentiation, and the trajectory of disease progression. With its capacity to inject a human touch into the AI-driven narrative, this academic-style prose aims to strike a balance between reflecting the sophistication of emerging biotechnology and lauding the quintessential human characteristic of scholarly curiosity.
4. Epigenetics and Gene Regulation:
Investigations into Small RNA sequencing play a crucial role in advancing our knowledge of epigenetic regulation and the control of gene expression. Through meticulous profiling of small RNA-induced epigenetic alterations namely, DNA methylation and histone modifications, it becomes possible to shed light on the complex interaction occurring between small RNAs and chromatin remodeling complexes. This, in turn, offers profound insights into the mechanisms underpinning gene silencing, as well as the inheritance of epigenetic traits. Altogether, such studies bring forward an enhanced understanding of the intricate layers of genetic regulation, masked by epigenetic modifications.
5. Evolutionary Studies and Comparative Genomics:
The employment of small RNA sequencing provides a robust platform for the progression of comparative genomic and evolutionary studies across a multitude of species. Through the detailed examination of the unique expression profiles of small RNAs across various organisms, it allows researchers to delve into the concurrent aspects of evolutionary conservation, divergence and the adaption of small RNA pathways. This innovative approach illuminates our understanding of the progression of gene regulation and the complex traits that distinguish one species from another.
6. Environmental Monitoring and Biomonitoring:
In the realm of environmental evaluation and biomonitoring, the utilization of Small RNA sequencing exhibits significant potential. The profiling of small RNA expression within environmental specimens, encompassing soil, water, and air, affords researchers an avenue to evaluate ecosystem health, identify the presence of environmental contaminants, and monitor the influences of environmental stress factors on biodiversity and the efficacy of ecosystem operation. The insightful data provided by such application makes considerable strides toward the understanding and preservation of environmental equilibrium.
7. Agriculture and Crop Improvement:
Leveraging small RNA sequencing is integral to agricultural research and efforts towards crop enhancement. The in-depth study of gene regulation in plants, mediated by small RNA, allows for the identification of major regulatory mechanisms involved in the response to stress factors, resistance to diseases, and improvements in crop yield. Such utilization paves the way for the generation of genetically fortified crops with heightened resilience and augmented productivity, reinforcing the adoption of innovative and sustainable practices in agriculture.
8. Diagnostic and Therapeutic Development:
As a pivotal technique in modern science, small RNA sequencing paves the way for personalized medical advancements. Specific small RNA signatures associated with various diseases, once identified and analyzed, can be harnessed to construct precise diagnostic tests for preemptive disorder detection and patient categorization. Furthermore, the exploration of small RNA-based treatments, encompassing elements such as miRNA imitations or inhibitors, present encouraging pathways for the evolution of tailored therapeutic strategies.
For some questions related to small rna sequencing, please refer to the "Small RNA Sequencing Q&A"
References:
- Seguin J, Rajeswaran R, Malpica-Lo ́pez N, et al. De Novo Reconstruction of Consensus Master Genomes of Plant RNA and DNA Viruses from siRNAs. Plos one. 2014, 9: e88513.
- Wu H, Li B, Iwakawa H, et al. Plant 22-nt siRNAs mediate translational repression and stress adaptation. Nature. 2020.
- Natsuko Izumi et al. Zucchini consensus motifs determine the mechanism of pre-piRNA production. Nature, 2020, doi:10.1038/s41586-020-1966-9.
- Li S, Xu ZP and Sheng JH. tRNA-derived small RNA: a novel regulatory small non-coding RNA. Genes. 2018, 9:246.
- Zhu L, Li J, Gong Y, Wu Q, et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol Cancer. 2019, 18(1):74.
- Benesova S, Kubista M, Valihrach L. Small RNA-Sequencing: Approaches and Considerations for miRNA Analysis. Diagnostics (Basel). 2021 May 27;11(6):964.
- Li, J., Kho, A.T., Chase, R.P. et al. COMPSRA: a COMprehensive Platform for Small RNA-Seq data Analysis. Sci Rep 10, 4552 (2020).
- Peter Androvic Small RNA-Sequencing for Analysis of Circulating miRNAs: Benchmark Study The Journal of Molecular Diagnostics 2022
- Dard-Dascot, C., Naquin, D., d'Aubenton-Carafa, Y. et al. Systematic comparison of small RNA library preparation protocols for next-generation sequencing. BMC Genomics 19, 118 (2018).
- Caroline Koch Highly multiplexed detection of microRNAs, proteins and small molecules using barcoded molecular probes and nanopore sequencing bioRxiv 2022


 Sample Submission Guidelines
Sample Submission Guidelines
