The Challenge and Workflow of Small RNA Sequencing
What Is Small RNA Sequencing?
Small RNA sequencing (Small RNA-Seq), facilitated by Next-Generation Sequencing (NGS) technology, is an instrumental technique for the isolation and acquisition of information about non-coding RNA molecules. This method allows the distinction and evaluation of small RNAs, including the discovery of novel variants and prediction of their prospective functions. By capitalizing on such technology, small RNAs can be differentiated from larger RNA families, thereby enhancing our understanding of their roles in cellular functions and gene expression.
Small RNA sequencing represents an increasingly popular approach to address the biological issues of miRNAs and other small non-coding RNAs (sncRNAs), such as piwi-interacting RNAs (piRNAs), small interfering RNAs (siRNAs), transcription initiation RNA (tiRNAs). MiRNAs, piRNAs, and siRNA are the three most focused sncRNAs and are widely studied by sequencing since they have been reported to play a vital role in post-translational regulation of gene expression. Small RNA sequencing has become a good standard for both small RNA discovery and small RNA profiling since they can sequence the entire complement of small RNA with high throughput and high sensitivity. Compared with microarrays and qPCR, small RNA sequencing doesn't require a priori knowledge of sequence. In addition to the acquirement of the complete range of miRNA and small RNA species, small RNA sequencing also helps understand how post-transcriptional regulation affect phenotype and identify novel biomarkers. It has been widely applied in cancer and complex disease research.
Small RNAs comprise a large category of regulatory molecules, including miRNA, ncRNA, siRNA, snoRNA, piRNA, rasiRNA, which are found ubiquitously across all biological entities. These small RNAs regulate organismal growth, development, and disease progression through various means such as mRNA degradation, translational repression, heterochromatin formation, and DNA elimination. Small RNA (sRNA) sequencing, generally undertaken on the Illumina HiSeq platform, facilitates comprehensive analysis of miRNAs, siRNAs, and piRNAs present in samples. Moreover, this technology can identify known sRNAs, predict novel ones, and anticipate sRNA target genes, thereby providing a powerful tool for examining small RNA functionality and regulatory mechanisms.
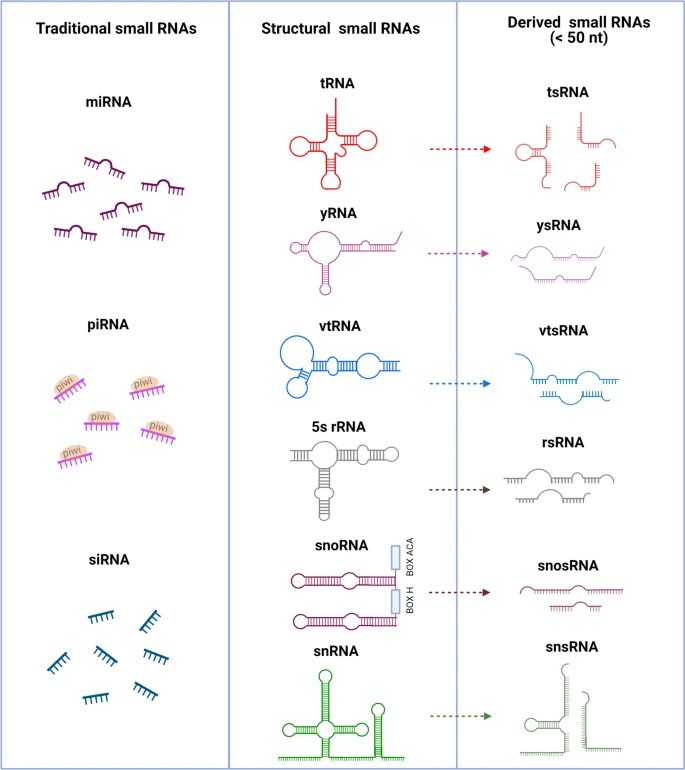 Figure 1. The structure and classification of small RNAs. (Xiong et al., 2023)
Figure 1. The structure and classification of small RNAs. (Xiong et al., 2023)
Services you may interested in
Advantages of Small RNA Sequencing
Small RNA sequencing is a high-throughput technique employed to investigate non-coding RNAs such as microRNAs (miRNA), siRNA, and piRNA, among others. This method enables the direct generation of miRNA sequencing libraries from total RNA, hence shedding light on the role of non-coding RNAs. Through this approach, we can not only understand how post-transcriptional regulation affects phenotypes but also identify novel biological markers and capture a comprehensive range of small RNA and miRNA species.
High-throughput nature: One sequencing run can yield over 5 million sequence reads.
High sensitivity: High-throughput sequencing of small RNAs holds an inherently high sensitivity, which can detect scarce non-coding RNA molecules. This feature enables researchers to identify and quantify low-abundance non-coding RNAs such as microRNAs.
Comprehensiveness: This methodology allows the detection and identification of all types of small RNA molecules present in a sample, including microRNAs, siRNAs, and piRNAs, among others. This level of comprehensiveness lets researchers thoroughly view the expressive landscape of non-coding RNAs.
High Resolution: Small RNA sequencing allows for the detection of minor variances in small RNA at the single base level; Precision: Highly accurate quantification ranging from few to several million copies. Including their length and structural characteristics, this high resolution enables researchers to delve into the function and regulatory mechanisms of non-coding RNA.
No Reliance on Known Information: It can identify known small RNAs, as well as uncover novel ones.
Excellent Reproducibility: In-depth sequencing guarantees the randomness of the samples, providing excellent reproducibility without the necessity for repeat experiments.
Data Diversity: The rich diversity of data generated by small RNA sequencing provides a wealth of information beneficial to biological research. The analysis of sequencing data enables the understanding of non-coding RNA expression patterns, their regulatory networks, and associated disease-related changes.
Cost-efficiency: With the advancement of sequencing technology, the costs associated with small RNA sequencing continue to decrease, rendering it financially accessible to an increasing number of labs and research projects. This cost reduction not only lowers the barrier to research but also promotes the broad application of non-coding RNA studies.
The Challenges of Small RNA Sequencing
Researchers have gradually found that the sequencing data containing differential expression of small RNAs is sometimes inconsistent with microarray, qPCR, and Northern blot results. This has been primarily attributed to the RNA-ligase-dependent bias for particular adapter sequences introduced during small RNA library construction. Studies have suggested that randomizing the adaptor sequences close to the ligation junction can reduce ligation bias and optimize sequencing results. Bioo Scientific's NEXTflex Small RNA-seq kit is currently the only kit commercially available that reduces ligation associated bias. This kit utilizes adapters with 4nt random ends which provides a more even coverage of individual small RNA species. A work by Baran-Gale et al., (2015) has suggested that the NEXTflex protocol is the least biased kit for small RNA sequencing.
Table 1. The commercially available kits for small RNA library construction.
| Kits | Matched sequencing platforms | Strategies for reducing bias |
| NEBNext Small RNA Library Prep Kit ((NEB) | Illumina platforms | Using polyethylene glycol (PEG) |
| NEXTflex Small RNA-seq kit V3 (BIOO Scientific) | Illumina platforms | Using both randomized adapters and PEG |
| SMARTer smRNA-Seq kit (Clontech) | Illumina platforms | Avoiding ligation altogether |
| CATS Small RNA-seq Kit (Diagenode) | Illumina platforms | Avoiding ligation altogether |
| TruSeq Small RNA Library Prep Kit (Illumina) | Illumina platforms | Indexes added during PCR |
The primary sources of bias in small RNA sequencing include ligation bias and PCR bias. Ligation bias stems from the differences in the secondary structure of miRNA and adapters during the ligation reaction, which leads to certain miRNAs having a higher ligation efficiency than others. This, in turn, impacts the accuracy of the data. Several strategies to mitigate this issue include improving adapter design to reduce bias, using computational corrections to adjust expression values, deploying specific enzymes to remove certain modifications, and increasing the randomness of the adapters.
Another source of bias is PCR bias, caused by the differing amplification efficiencies of molecules of various lengths and secondary structures during the PCR process. To alleviate this bias, molecular barcodes could be used to label PCR products, thereby distinguishing identical molecules that have been amplified via PCR. The final step in the small RNA sequencing workflow is sequencing. This step might be associated with certain types of bias (such as lane effects and pooling effects), but these biases are generally considered as secondary biases that subtly influence the overall level of bias.
Currently, there are primarily three methods to address different bias issues: (1) original two-adapter ligation; (2) improved dual-adapter ligation; (3) ligation-free (adenylation-based) methods. Moreover, a suite of alternative technologies reliant on the hybridization of miRNA and target probes wholly avoids the steps of ligation or polyadenylation, and in certain cases, also eliminates the PCR phase and NGS reads. These techniques usually offer simplified workflow suitable for routine and automated analysis. However, their analyzable miRNA spectrum, determined by a set of pre-designed probes, is limited, thereby constraining their potential for discovery.
(1) The original two adapter ligation methods
The traditional method of dual-adapter ligation involves initially connecting a pre-adenylated 3' adapter, followed by the 5' adapter (Fig.2). This approach is favoured for its applicability across a broad spectrum of small RNA types, its ability to detect RNAs with particular modifications, and its long-standing historical use. However, inherent limitations exist. Adapter ligation bias may lead to certain miRNAs being overestimated or underestimated, or potentially evading detection altogether. The formation of adapter-dimers may compromise read counts, further affecting data precision. Additionally, this method falls short of capturing specific types of small RNA, such as snRNAs with 3' end modifications.
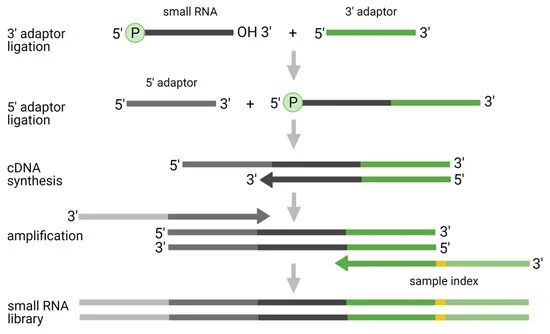 Figure 2. Original two-adaptor ligation protocol for small RNA-seq analysis. (Benesova et al., 2021)
Figure 2. Original two-adaptor ligation protocol for small RNA-seq analysis. (Benesova et al., 2021)
(2) The improved method based on two adapter ligation
Improved dual-adapter ligation methods aim to mitigate or counteract ligation and PCR biases. At present, three techniques are available: (i) coupling both adapters with random nucleotides (Figure 3A); (ii) ligation of a single adapter followed by circularization (Figure 3B); and (iii) the integration of molecular barcodes with a dual adapter. Random adapters reduce ligation bias through inclusion of random sections, which may require careful management to evade the impacts of PCR bias. The approach of single-adapter ligation and subsequent circularization leverages intramolecular circularization to lower ligation bias, although there may still be potential 3' end biases. In contrast, the method involving molecular barcodes mitigates PCR bias by incorporating molecular barcodes.
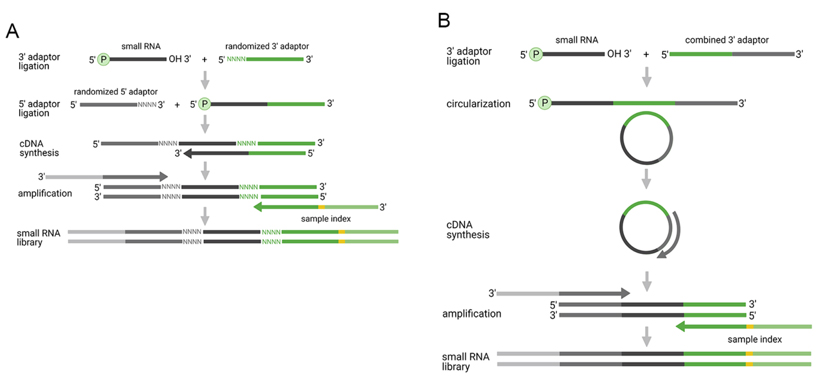 Figure 3. Improved two-adaptor ligation-based methods for small RNA-seq analysis. The reduction of ligation or PCR bias is achieved by: (A) randomized adaptors; (B) single adaptor-ligation and circularization; and (C) molecular barcodes. (Benesova et al., 2021)
Figure 3. Improved two-adaptor ligation-based methods for small RNA-seq analysis. The reduction of ligation or PCR bias is achieved by: (A) randomized adaptors; (B) single adaptor-ligation and circularization; and (C) molecular barcodes. (Benesova et al., 2021)
(3) Ligation-free method
The ligation-free approaches primarily include schemes based on polyadenylation and template switching (Fig. 4), as well as probe-based technologies, all of which enable targeted analysis of known miRNAs. These techniques offer a streamlined workflow and computational analysis while circumventing the biases associated with linking and PCR steps. The approach relying on polyadenylation and template switching makes use of reverse transcriptase (RTase) template-switching activity to add 5' end adapters, as opposed to traditional ligation methods. This method's independence from ligation allows for a more accurate quantification of miRNAs, sidestepping biases introduced by ligation steps. However, it does have certain disadvantages, including some reads not aligning with miRNA sequences, leading to lower miRNA mapping rates and potential increases in sequencing costs.
Probe-based technologies employ specific probes for the targeted analysis of known miRNAs. For instance, the Nanostring nCounter uses direct molecular barcodes and color-coded probes for digital detection of miRNAs, making it suitable for the quantification of over 800 different miRNAs. The FirePlex miRNA assay combines hydrogel nanoparticle technology with miRNA-specific regions and universal adapters, allowing the quantification of 65 miRNA species in a single reaction and eliminating deviation introduced by the separation process. Meanwhile, EdgeSeq utilizes probes that bind specifically for miRNA analysis and sequencing. Its library preparation process is automated, and the computational analysis standardized, thus yielding comparable and reliable outcomes.
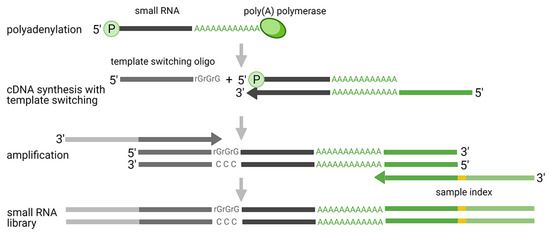 Figure 4. Polyadenylation and template switching mechanism applied in small RNA-seq analysis. (Benesova et al., 2021)
Figure 4. Polyadenylation and template switching mechanism applied in small RNA-seq analysis. (Benesova et al., 2021)
The Workflow of Small RNA Sequencing
The workflow of small RNA sequencing generally includes total RNA isolation, small RNA library construction, deep sequencing, and bioinformatics analysis.
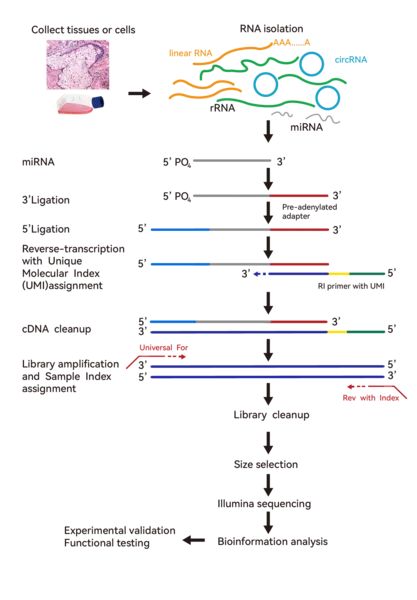 Figure 5. Small RNA sequencing technology pathway.
Figure 5. Small RNA sequencing technology pathway.
Enrichment of Small RNA
Given the characteristically short lengths of small RNAs, typically spanning between 20 to 30 nucleotides (nt), it necessitates the enrichment of the total RNA extracted. This process can be accomplished either through the utilization of a specific small RNA enrichment reagent kit, such as the miRNeasy Mini Kit, or via size-selective gel electrophoresis. The samples used for enrichment can range from cellular materials, tissues, to various body fluids.
Small RNA library construction
The 5' phosphate and 3' hydroxyl groups on the small RNA allow ligases to selectively target and capture these small RNA species. For small RNA sequencing, library construction typically starts with the ligated of pre-adenylated DNA adaptor to the 3'-end of the sRNAs by using a truncated version of T4 RNA ligase 2, following by the ligation of an RNA adaptor to their 5'-end using T4 RNA ligase 1. After this ligation, the products can be easily reversed transcribed into cDNA and amplified by PCR before high-throughput sequencing. There are many commercially available kits that generate small RNA libraries directly from total RNA sample (Table 1). The products are suitable for Illumina sequencing. The general principles of small RNA library construction are outlined in Figure 6, but different kits may have some different protocols. The process of gel purification is dedicated to quality control and size selection. According to your research interests, the appropriate size of sequences is recovered by bead-based size selection or PAGE purification for deep sequencing. 18-40 bp insert cDNA library is a typical choice which covers miRNA, siRNA, and piRNA. Please note that sequencing miRNA and mRNA require two separate library protocols, even with the same total RNA sample. After the quality control of small RNA libraries, high-throughput sequencing is then performed. Illumina platforms are the most popular instruments for small RNA sequencing, such as MiSeq and HiSeq.
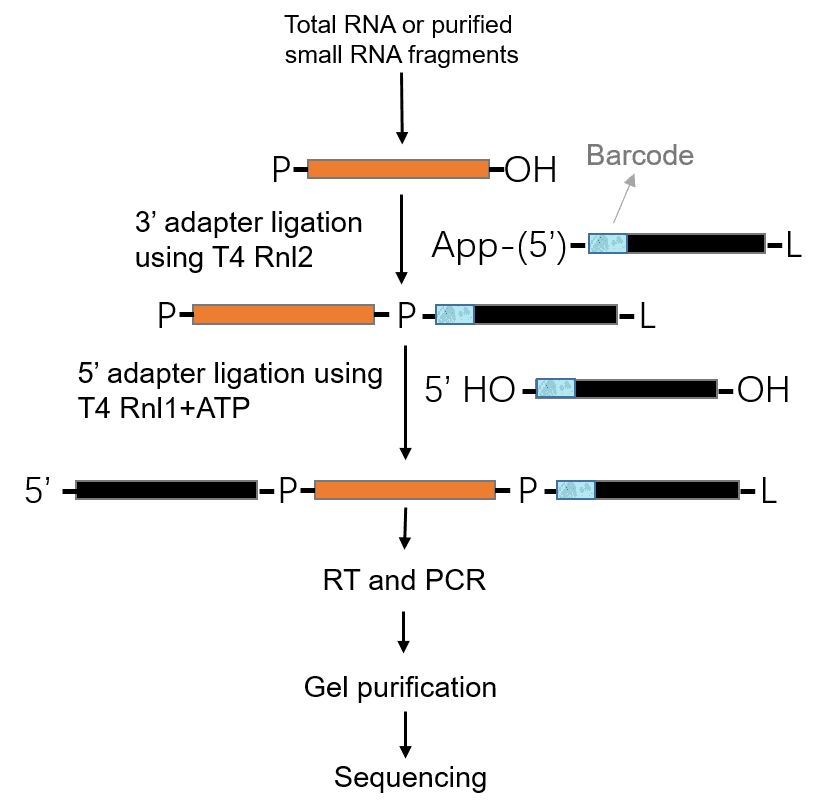 Figure 6. The principles of small RNA library construction.
Figure 6. The principles of small RNA library construction.
Bioinformatics analysis
Small RNA sequencing can be used for small RNA clustering, novel small RNA discovery, miRNA target prediction, differential expression of small RNA, evolutionary analysis, and functional analysis. But prior to these, raw sequencing data need to be preprocessed and normalized. The data preprocessing involves removal of adapter and barcode, size selection, removal of complex reads, and the generation of unique reads. Normalization is the process to make expression levels comparable across libraries. Rfam and miRBase are two common databases for small RNA analysis. Rfam is an open-access database providing information about tRNA, rRNA, and snoRNA, etc. miRBase contains sequences and annotations of all known miRNAs across species.
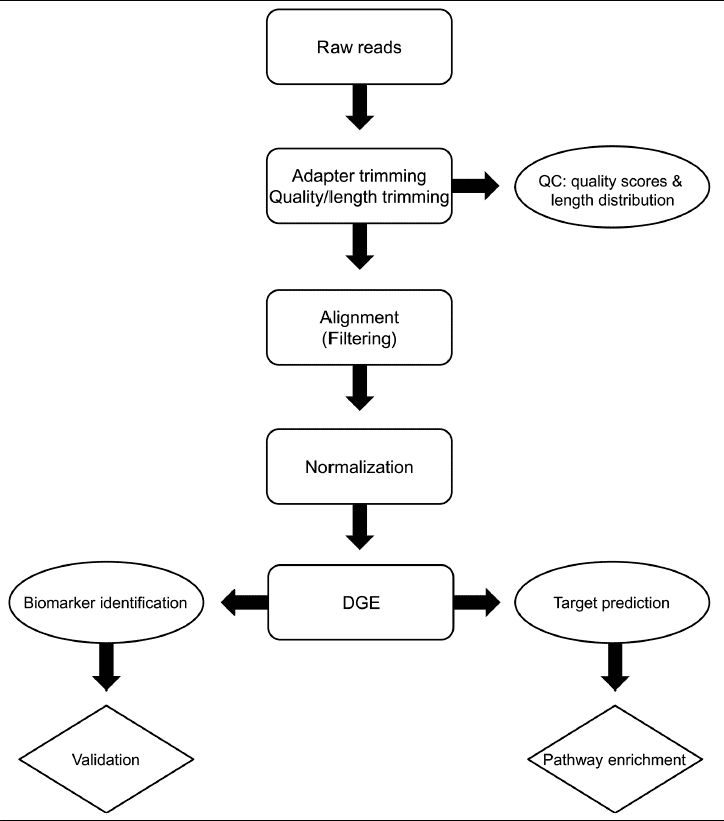 Figure 7. The bioinformatics analysis workflow of small RNA sequencing data (Buschmann et al. 2016).
Figure 7. The bioinformatics analysis workflow of small RNA sequencing data (Buschmann et al. 2016).
Data Quality Control: The reliability of information analysis results hinges upon the quality of sequencing data. Therefore, the raw sRNA sequencing data undergoes stringent quality control processes including adapter trimming, removal of sequences containing N bases, filtering out low-quality bases, and length selection. This ensures the acquisition of high-quality sequencing data, while simultaneously facilitating the enumeration of small RNA species and characterization of their length distribution.
Reference Genome Alignment: Starting from the Clean reads obtained after length selection from each sample, alignment to the reference genome is performed, followed by statistical analysis of alignment results.
Known miRNA Analysis: Alignment of reads to the miRNA database enables the identification of known microRNAs.
ncRNA Analysis: Alignment of reads to species-specific ncRNA sequences or the Rfam database facilitates the identification of rRNA, tRNA, snRNA, and snoRNA.
Novel miRNA Prediction: Utilizing miREvo for novel miRNA prediction, accompanied by analyses of nucleotide preference and secondary structure.
Gene Expression and Differential Analysis: Quantification of known and novel miRNAs based on alignment results, followed by differential miRNA analysis to elucidate differential expression among samples or groups.
miRNA Target Gene Prediction: Leveraging both miRanda and RNAhybrid software tools for the prediction of differential miRNA target genes.
Functional Enrichment: Utilizing functional annotation databases such as GO and KEGG, significant enrichment of functional information pertaining to differentially expressed miRNA target genes among samples or groups is obtained.
Application of Small RNA Sequencing
Small RNA sequencing is a pivotal technique in biology, widely utilized across various research domains, particularly in gene regulation, disease diagnosis, and biomedical investigation.
miRNA Expression Analysis:
MicroRNAs (miRNAs) are essential non-coding RNAs implicated in gene expression regulation. Small RNA sequencing enables comprehensive and high-throughput assessment of miRNA expression levels, thereby elucidating the functional and regulatory networks of miRNAs within organisms.
miRNA Functional Research:
Utilizing small RNA sequencing enables the identification of miRNA target genes and further exploration of the mutual interactions between miRNAs and their targets. This approach delves deeply into unraveling the biological functions and regulatory mechanisms of miRNAs.
Biomarker Discovery:
Given the pivotal roles of miRNAs in the onset and progression of numerous diseases, small RNA sequencing serves as a potent tool for discovering disease-associated miRNAs. Consequently, it offers potential biomarkers for disease diagnosis and therapeutic interventions.
Investigating miRNA-disease Associations:
By juxtaposing miRNA expression profiles under both healthy and diseased states, it is possible to discern disease-associated miRNAs. This unveiling could provide significant insights into elucidating disease pathogenesis and engendering innovative treatment methodologies.
Research on miRNA-drug Interactions:
Small RNA sequencing can facilitate the exploration of interactions between miRNAs and drugs. This includes the influence of pharmaceutical compounds on miRNA expression, as well as the regulatory role of miRNAs on drug metabolism and therapeutic efficacy.
Investigations into Biological Evolution:
The comparative analysis of miRNA expression profiles between different species or individuals through small RNA sequencing can spotlight miRNA's conservativeness and diversity in the evolutionary process of organisms.
Research on Environmental Stress Response:
Given the critical role of miRNA in an organism's response to environmental stress, employing small RNA sequencing provides a method to study how said stress influences the miRNA expression profile, thereby probing the underlying mechanisms of biological adaptation.
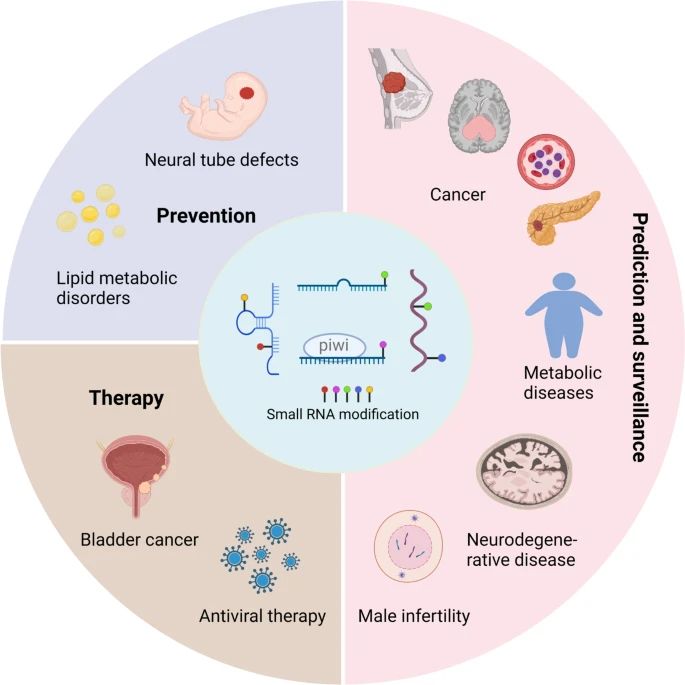 Figure 8. Applications of small RNA modifications. (Xiong et al., 2023)
Figure 8. Applications of small RNA modifications. (Xiong et al., 2023)
For more detailed bioinformatics pipeline for small RNA sequencing, please refer to this page. In addition to small RNA sequencing, we further provide other transcriptomic sequencing services, including RNA-seq, bacterial RNA sequencing, lncRNA sequencing, circRNA sequencing and degradome sequencing.
References:
- Baran-Gale J, Kurtz C L, Erdos M R, et al. Addressing Bias in Small RNA Library Preparation for Sequencing: A New Protocol Recovers MicroRNAs that Evade Capture by Current Methods. Frontiers in Genetics, 2015, 6(e73240).
- Buschmann D, Haberberger A, Kirchner B, et al. Toward reliable biomarker signatures in the age of liquid biopsies-how to standardize the small RNA-Seq workflow. Nucleic acids research, 2016, 44(13): 5995-6018.
- Fuchs R T, Sun Z, Zhuang F, et al. Bias in ligation-based small RNA sequencing library construction is determined by adaptor and RNA structure. Plos One, 2015, 10(5): e0126049.
- Benesova S, Kubista M, Valihrach L. Small RNA-sequencing: approaches and considerations for miRNA analysis. Diagnostics, 2021, 11(6): 964.
- Xiong Q, Zhang Y. Small RNA modifications: regulatory molecules and potential applications. Journal of Hematology & Oncology, 2023, 16(1): 64.


 Sample Submission Guidelines
Sample Submission Guidelines
