Applications of Low-Pass Whole Genome Sequencing in Clinical Cytogenetics
Clinical cytogenetics is a subject that studies the structural and functional abnormalities of chromosomes and their effects on human health. Traditional chromosome analysis methods, such as routine karyotype analysis and fluorescence in situ hybridization (FISH), are still effective in some cases, but they have some limitations, such as low resolution and limited detection range. With the development of genomics technology, whole genome sequencing (WGS) and low-pass whole genome sequencing (LP-WGS) have gradually become important tools in clinical cytogenetics.
Introduction to LP-WGS in Clinical Cytogenetics
The application of LP-WGS in clinical cytogenetics mainly involves its potential in evaluating the contribution of low-frequency and common genetic variations to human disease-related quantitative phenotypes. By low-depth sequencing covering the whole genome, this technology can identify the genetic variation that affects gene expression and biomarker phenotype, thus providing important information for the diagnosis and treatment of diseases. LP-WGS can effectively identify genetic variations that affect gene expression and biomarker phenotype, providing a new perspective for understanding the genetic basis of complex diseases. It is of great significance in evaluating the risk of carriers of rare single gene mutation, which is helpful to estimate the disease risk of individuals more accurately and guide clinical decision-making.
With the continuous improvement of technology and further reduction of cost, LP-WGS is expected to play a greater role in clinical cytogenetics. For example, some researchers has verified the effectiveness of LP-WGS in prenatal diagnosis, and believes that it has broad application prospects in the future. The application background of LP-WGS technology in clinical cytogenetics is mainly reflected in its high resolution, low cost and wide clinical applicability. These characteristics make it an effective supplement to traditional chromosome analysis methods, and provide new tools and methods for genomics research and clinical diagnosis.
Overview of Clinical Cytogenetics and LP-WGS
Clinical cytogenetics plays a vital role in modern medicine, especially in the diagnosis and treatment of hereditary diseases, cancer and other complex diseases. Its main functions include the following parts.
Disease diagnosis: Clinical cytogenetics can diagnose many hereditary diseases by detecting chromosome abnormality and copy number variation (CNVs). For example, chromosome microarray analysis (CMA) and WGS have become common methods to diagnose unexplained diseases such as developmental delay, mental retardation and autism spectrum disorder. In prenatal diagnosis, LP-WGS has been proved to be an effective tool to detect fetal chromosomal abnormalities, such as simple tetralogy of Fallot.
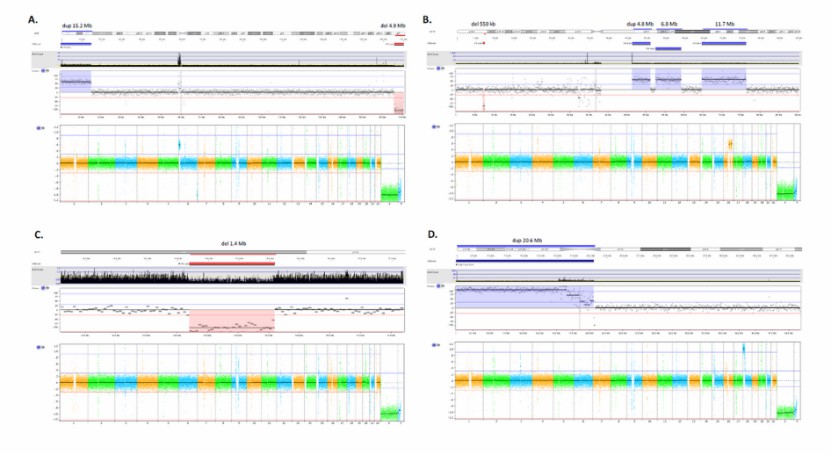 Examples of CNVs detected by LP-WGS (Mazzonetto et al., 2024)
Examples of CNVs detected by LP-WGS (Mazzonetto et al., 2024)
Disease classification and management: By identifying specific gene variants and CNVs, clinical cytogenetics helps to classify diseases more accurately, thus guiding the choice of treatment schemes. For example, low-pass genome-wide sequencing has been proved to be an effective diagnostic tool in patients with neurodevelopmental disorders and congenital abnormalities. In cancer research, the detection of CNVs is very important for understanding the genetic background of tumors and choosing appropriate treatment strategies.
Genetic counseling and support: Clinical cytogenetics not only provides diagnostic information, but also provides genetic counseling for patients and their families to help them understand the genetic risks and possible genetic patterns of diseases. By interpreting the test results, clinical cytogenetics helps patients and their families to make informed medical decisions and provide continuous psychosocial support.
LP-WGS plays an important role in the detection of chromosome abnormality and CNVs. On the one hand, by analyzing the coverage of sequencing reads, we can detect chromosome number and structural abnormality from the genome-wide level, and we can find tiny and hidden abnormalities. On the other hand, it can detect the whole genome CNVs without bias, accurately and quantitatively analyze the change degree of copy number, and also make functional annotation and analysis combined with clinical phenotype to determine the relationship between CNVs and diseases, which provides an important basis for disease diagnosis, genetic counseling and personalized medical care.
High sensitivity and specificity: LP-WGS technology can detect tiny CNVs that cannot be found by traditional karyotype analysis. For example, studies have shown that low-pass genome-wide sequencing has obvious advantages in detecting tiny CNVs, which can make up for the lack of resolution in karyotype analysis. In prenatal diagnosis, the detection rate of CNVs detected by LP-WGS was 18.85%, of which pathogenic CNVs accounted for 91.89%. This shows that LP-WGS has high sensitivity and specificity in detecting pathogenic CNVs.
Wide range of applications: LP-WGS technology is not only suitable for the detection of fetal chromosomal abnormalities, but also widely used in clinical scenarios such as spontaneous abortion and recurrent abortion. For example, in spontaneous abortion tissues, the detection rate of CNVs detected by high-throughput sequencing technique (including LP-WGS) was 42.92%. In the prenatal diagnosis of high-risk fetus, LP-WGS combined with karyotype analysis can improve the detection rate of abnormal chromosomes, which is of great significance to prenatal diagnosis.
Combined application with other technologies: LP-WGS is often combined with chromosome karyotype analysis and chromosome microarray analysis (CMA) to improve the accuracy and comprehensiveness of detection. For example, the combination of low-pass whole genome sequencing and karyotype analysis can detect more tiny CNVs and improve the accuracy of prenatal diagnosis. In some studies, the combination of LP-WGS and chromosome microarray analysis can detect fetal chromosomal abnormalities more effectively, especially in detecting tiny CNVs.
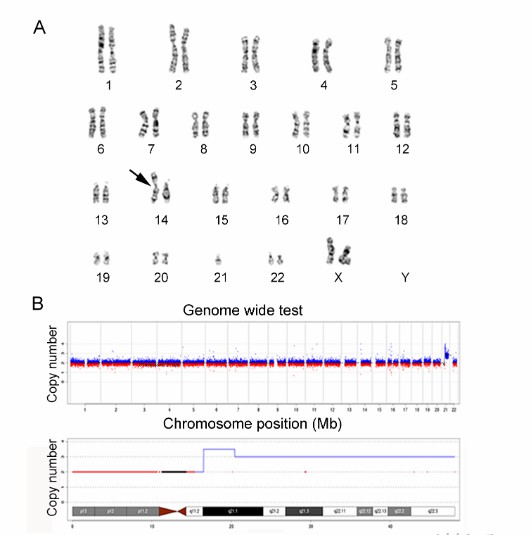 Karyotyping and LP-WGS results of case (Zhang et al., 2022)
Karyotyping and LP-WGS results of case (Zhang et al., 2022)
The role of LP-WGS in the detection of chromosome abnormality and copy number variation is mainly reflected in its high sensitivity, wide application range, joint application with other technologies, significant clinical application value and technical advantages. These characteristics make it an indispensable tool in modern genetic research and clinical diagnosis.
Services you may interested in
Want to know more about the details of LP-WGS? Check out these articles:
Applications of LP-WGS in Clinical Cytogenetics
LP-WGS is a technology to reduce the cost and computational requirements by reducing the sequencing depth, while still detecting chromosome abnormalities and CNVs. The following are its specific application methods and principles:
Technical principle
- Coverage: LP-WGS usually uses low coverage (such as 1× or 2×), which means that each base is sequenced once or twice on average. This low coverage can significantly reduce the sequencing cost, but it can still detect large CNVs and some types of structural variation (SV).
- Data processing: The sequencing data is processed by bioinformatics tools, including reading segment comparison, normalization and boxing. Commonly used tools include CNVKit, GATK, etc. These tools can identify copy number variation and other types of genetic variation.
 CNVs analysis by LP-WGS using the CNVkit software (Olivier et al., 2024)
CNVs analysis by LP-WGS using the CNVkit software (Olivier et al., 2024)
Detection method
- SNP chip replacement: LP-WGS can be used as an alternative technology for chromosome microarray (CMA) to detect copy number variation. This method has advantages in cost and time.
- Reference standard: By comparing with the known SNP array data, verify the detection results of LP-WGS. For example, in the diagnosis of hematological tumors, LP-WGS is highly consistent with the detection results of SNP array data.
Clinical application
- Prenatal diagnosis: LP-WGS sequencing can be used for prenatal diagnosis, and detect chromosomal abnormalities and CNVs of the fetus. For example, in a study of Hubei Maternal and Child Health Hospital, 24 fetuses with simple tetralogy of Fallot were detected by low-pass whole genome sequencing, and 5 cases were found to have CNVs.
- Cancer diagnosis: In cancer diagnosis, LP-WGS can detect CNVs in tumor cells, providing important information for early detection and treatment. For example, in patients with bladder cancer, the CNVs detected by LP-WGS is highly correlated with clinical manifestations.
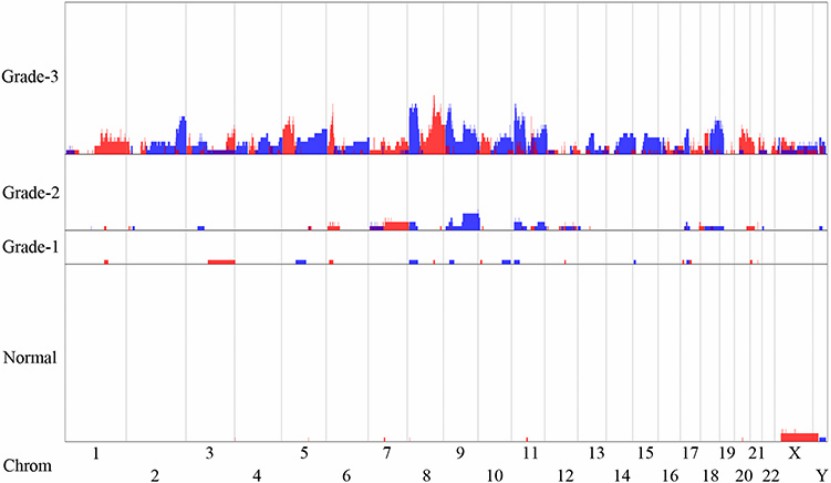 CNVs in urine sediment of bladder tumors (Cai et al., 2021)
CNVs in urine sediment of bladder tumors (Cai et al., 2021)
- Diagnosis of rare diseases: LP-WGS also shows potential in the diagnosis of rare diseases. For example, the CNVs detected by LP-WGS can be used to diagnose some complex genetic diseases.
Case Studies
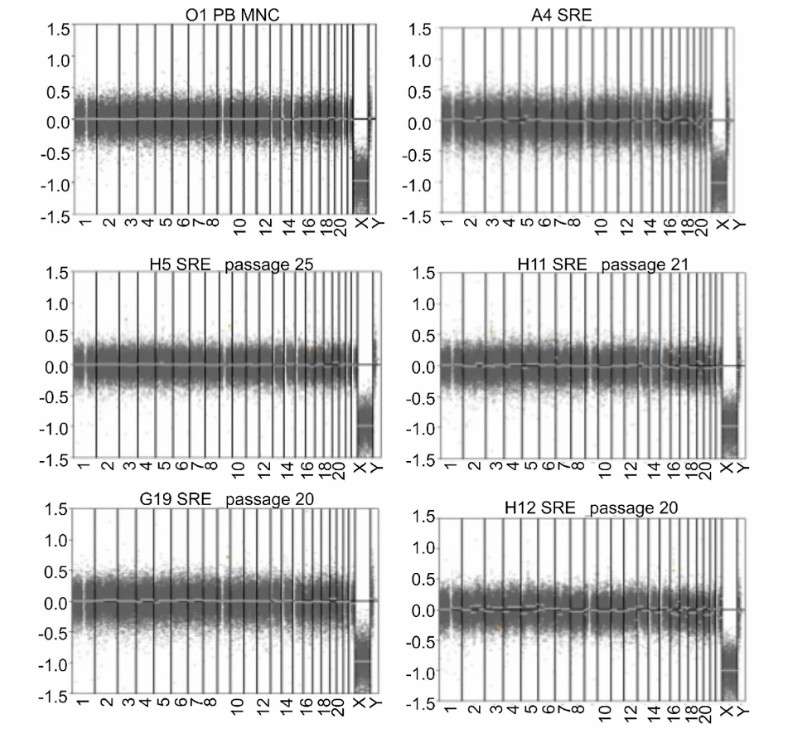
The application of LP-WGS in clinic has been widely verified, especially in preimplantation genetic screening (PGT-A) and preimplantation genetic diagnosis (PGT-G). According to the research report published on January 1st, 2024, eight common Coriell cell lines were used to screen the chromosome aneuploidy of PGT-A, covering the common types of chromosome abnormalities in PGT-A, including normal chromosome types, autosomal abnormalities and sex chromosome abnormalities. The results showed that the successful amplification and accuracy were over 99.9% by LP-WGA and low-magnification sequencing (LM-WGS).
In PGT-G, the research team developed the laboratory protocol LP-WGA method. Compared with genomic DNA, the quality of DNA produced by LP-WGA is poor, but the sensitivity and specificity are equivalent. Compared with the reference data of National Institute of Standards and Technology (NIST), the total accuracy and specificity of LP-WGA at the genome level reached 99.99%. In addition, the accuracy of embryo biopsy was also verified, and the overall accuracy and specificity were 96.6%, 99.6%, 99.8% and 98.1% respectively.
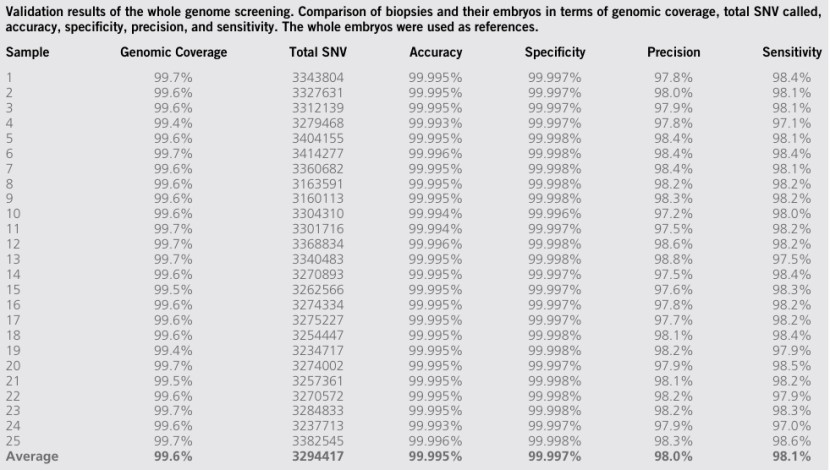 Validation results of the LP-WGS (Xia et al., 2024)
Validation results of the LP-WGS (Xia et al., 2024)
The application effect of LP-WGS in clinical cytogenetics is remarkable, especially in single cell analysis and cancer research. The development of digital microfluidic technology provides a new solution for single cell analysis. The fully automatic single cell processing platform based on digital microfluidic can realize efficient, fast and nondestructive fully automatic single cell separation, and has good universality, robustness and biocompatibility. The platform can realize rapid, automatic and fully integrated preparation of single-cell genome-wide amplification and sequencing samples, and solve the problem of insufficient high-throughput sequencing coverage caused by amplification bias in traditional methods.
In cancer research, genome-wide association studies (GWAS) is widely used to identify genetic variation related to diseases. For example, studies have found the regulatory role of HOXB13 gene in prostate cancer. By sequencing the whole exons of prostate cancer patients, the researchers screened out the risk mutation sites related to HOXB13 gene, and evaluated the expression differences of different alleles by ECL algorithm and eQTL analysis. These studies provide important clues for understanding the function of cancer risk mutation sites.
The application of LP-WGS in clinical cytogenetics not only improves the accuracy and reliability of detection, but also provides a new technical means for single cell analysis and cancer research. These advances provide important scientific basis and technical support for clinical diagnosis and treatment.
Advantages and Challenges
LP-WGS has many advantages in clinic. It can simultaneously detect chromosome number and structural abnormalities at the genome-wide level, and is more sensitive to small or chimeric abnormalities. It can also detect genome-wide CNVs without bias, find new unknown CNVs, accurately quantify CNVs by accurately calculating the depth and coverage of sequencing reads, and make functional annotation and analysis in combination with clinical phenotypes to determine the relationship between CNVs and diseases, so as to diagnose, classify and diagnose diseases.
The advantages of LP-WGS
- High coverage and high accuracy: LP-WGS can achieve high coverage through low sequencing depth (usually 10x or less), and can detect most common mutations and some rare mutations. This high coverage and high accuracy make it have obvious advantages in clinical diagnosis, especially in the diagnosis of complex genetic diseases.
- Quick diagnosis: LP-WGS can complete sequencing and data analysis in a short time, which is especially important for clinical situations that need rapid diagnosis. For example, in the diagnosis of Phelan-McDermid syndrome, low-pass whole genome sequencing can quickly identify new missing regions and provide timely diagnosis and treatment suggestions for patients.
- Wide applicability: LP-WGS is suitable for many types of genetic diseases, including single-gene genetic diseases, chromosome abnormalities and complex genetic diseases. For example, in prenatal genetic diagnosis of fetal congenital heart disease, LP-WGS can detect CNVs, SNV and other mutations, providing important genetic information for clinic.
 Chromosomes karyotype result (Lei et al., 2016)
Chromosomes karyotype result (Lei et al., 2016)
- Economical and efficient: Compared with high-throughput sequencing, LP-WGS has lower cost and is suitable for large-scale clinical application. Its low cost and high speed make it have high practical value in areas with limited resources.
- Multiomics integration analysis: LP-WGS can be combined with other omics data (such as transcriptomics, protein omics, etc.) for multi-omics integration analysis, so as to further improve the accuracy and comprehensiveness of diagnosis. For example, in the study of prostate cancer, by integrating genome, transcriptome and protein data, we can understand the molecular mechanism of tumor more comprehensively.
The challenges of LP-WGS
- Sensitivity and specificity of mutation detection: Although LP-WGS can detect most common mutations, its sensitivity to rare mutations and low-frequency mutations is low. In order to solve this problem, we can increase the sequencing depth or adopt more advanced bioinformatics methods to improve the detection sensitivity.
- Complexity of data interpretation: A large amount of data generated by low-pass whole genome sequencing requires complex bioinformatics analysis to interpret. Therefore, developing efficient bioinformatics tools and algorithms is the key. For example, using the mutation detection method based on deep learning can improve the accuracy and efficiency of data interpretation.
- Requirements for sample quality and quantity: LP-WGS requires the quality and quantity of samples, especially in high-throughput sequencing, the purity and DNA quality of samples will affect the sequencing results. Adopting high-quality samples and optimized sample processing methods can effectively solve this problem.
- Ethical and privacy issues: Privacy protection and ethical issues of genomic data are particularly important in clinical application. It is necessary to establish strict ethical review mechanism and data protection measures to ensure the safety and privacy of patient information.
- Technical standardization and quality control: Standardization and quality control of low-pass whole genome sequencing technology are the key to ensure the reliability of clinical application. The reliability and consistency of sequencing results can be improved by establishing a unified technical standard and quality control process.
LP-WGS has obvious advantages in clinic, but it also faces some challenges. By continuously optimizing technology, improving bioinformatics methods and strengthening ethical management, the clinical application effect can be further improved.
Conclusion
LP-WGS is very important in clinical cytogenetics. It can detect chromosome number and structural abnormalities in the whole genome, and it is more sensitive to tiny and chimeric abnormalities. It can detect CNVs without bias, and make functional annotation and analysis based on clinical phenotype and other information, so as to determine the correlation between CNVs and diseases, provide basis for disease diagnosis, classification and prognosis evaluation, help find therapeutic targets, and promote the development of personalized medicine, which is of great significance to disease diagnosis, genetic mechanism research and precision medical practice in clinical cytogenetics.
LP-WGS has great potential in improving detection efficiency and accuracy. Technological development will speed up sequencing, enable clinicians to obtain results quickly and strive for treatment time for critical patients. At the same time, the optimization of sequencing instruments and algorithms can make the detection accuracy higher, identify tiny variation more accurately and reduce the rate of misdiagnosis and missed diagnosis.
In the future, LP-WGS will expand the scope of disease diagnosis, help the diagnosis of rare and complex diseases, promote personalized medical care, provide a basis for accurate medication and treatment plan formulation, and strengthen prenatal and neonatal screening. It will also deeply analyze the disease mechanism through the integration of multi-omics, realize remote diagnosis and tap the disease law by combining telemedicine and big data sharing, and push clinical diagnosis to a new height in an all-round way.
References:
- Mazzonetto PC, Villela D., et al. "Low-pass whole genome sequencing as a cost-effective alternative to chromosomal microarray analysis for low- and middle-income countries." Am J Med Genet A (2024): e63802 https://doi.org/10.1002/ajmg.a.63802
- Zhang S, Xu Y., et al. "Combined use of karyotyping and copy number variation sequencing technology in prenatal diagnosis." PeerJ (2022): e14400 https://doi.org/10.7717/peerj.14400
- Olivier E, Zhang S., et al. "Stem cell factor and erythropoietin-independent production of cultured reticulocytes." Haematologica (2024): 3705-3720 https://doi.org/10.3324/haematol.2023.284427
- Cai YX, Yang X., et al. "Low-Coverage Sequencing of Urine Sediment DNA for Detection of Copy Number Aberrations in Bladder Cancer." Cancer Manag Res (2021): 1943-1953 https://doi.org/10.2147/cmar.s295675
- Xia Y, Katz M., et al. "The first clinical validation of whole-genome screening on standard trophectoderm biopsies of preimplantation embryos." F S Rep (2024): 63-71 https://doi.org/10.1016/j.xfre.2024.01.001
- Lei D, Li S., et al. "Clinical and genomic evaluation of a Chinese patient with a novel deletion associated with Phelan-McDermid syndrome." Oncotarget (2016): 80327-80335 https://doi.org/10.18632/oncotarget.12552


 Sample Submission Guidelines
Sample Submission Guidelines
