The Significance of Integrated Analysis of ATAC-seq and RNA-seq
In the sphere of molecular biology, the integration of Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) and RNA sequencing (RNA-seq) technology brings profound understanding to the subtly complex mechanisms involved in genomic regulation. The ATAC-seq technique uses the Tn5 transposase to pinpoint accessible regions of chromatin, isolate highlighted DNA fragments, and carry out polymerase chain reaction (PCR) amplification prior to sequencing. Analysis of the resultant sequencing reads enables predictions about neighboring DNA regions, along with the recognition of transcription factor binding sites and the determination of nucleosome positioning within particular loci. This holistic strategy not only expands our knowledge of the regulatory genomic landscape but also underlines the capacity of human-conceived and executed methods in decoding the complexities of gene expression control.
Transcriptome sequencing stands as a ubiquitous tool for investigating gene expression dynamics. It is well-established that gene expression necessitates the binding of the Transcription Preinitiation Complex (PIC) to DNA, thereby completing the transcription process. Regions undergoing transcriptional activity are inherently open, elucidating the reason behind the prevalence of gene activity on euchromatic regions in contrast to gene silencing on heterochromatic regions of the chromosomes.
Integrated Analysis of ATAC-seq and RNA-seq
The combined experimental approach employing ATAC-seq and RNA-seq serves a dual purpose in elucidating the differential expression of genes attributed to open chromatin alterations. On one hand, it expounds upon genes undergoing significant expression changes due to these open chromatin modifications. On the other hand, by discerning distinct open chromatin regions, it facilitates a more precise prediction of transcription factors that exert regulatory control over target genes. This nuanced approach offers valuable insights into the regulatory mechanisms governing the target genes. The intricacies of life regulation arising from the diverse repertoire of transcription factors contribute to the complexity of the regulatory landscape. In light of the diverse transcription factors influencing life regulation, the relationship between chromatin accessibility and gene regulation has emerged as a focal point of interest among scientific researchers.
CD Genomics offers comprehensive services in library sequencing analysis, expediting delivery timelines. Our repertoire includes a diverse array of personalized analyses, numbering in the dozens, accompanied by individualized technical support.
Epigenomics
ATAC-Seq
Transcriptomics
Transcriptomic Data Analysis
Epigenomics Data Analysis Service
Learn more from resource
ATAC-Seq – A Method to Study Open Chromatin
How to Plan Your Next RNA Sequencing Experiment
Applications of RNA-Seq
Application of Integrated ATAC-seq and RNA-seq Analysis
| Application Area | Focus |
| Plant Research | Chromatin Accessibility Profiling |
| Identification of cis-Regulatory Elements | |
| Plant Development (Flowering, Root Development, Reproductive Transformation, Vernalization, etc.) | |
| Metabolite Synthesis | |
| Abiotic Stress Responses (Salt-alkali, High Temperature, Low Temperature, Frost Damage, Drought, Flooding, etc.) | |
| Biotic Stress Responses (Disease and Pest Infestations, Viral Infections, etc.) | |
| Animal Research | Identification of cis-Regulatory Elements |
| Tissue or Organ Specificity | |
| Cellular Development (Embryonic Development, Stem Cell Differentiation, etc.) | |
| Mechanisms of Disease Occurrence (Tumor Formation, Functional Study of Pathogenic Genes, etc.) | |
| Drug Mechanisms (Drug Responsiveness, Mechanisms of Drug Resistance, etc.) | |
| Viral Infection Mechanisms (Host Responses, etc.) | |
Case Study of Integrated ATAC-seq and RNA-seq Analysis
Integrative ATAC-seq and RNA-seq Analysis of the Longissimus Muscle of Luchuan and Duroc Pigs
Published: 2021
Journal: Frontiers in Nutrition (Impact Factor: 6.57)
In the domain of skeletal muscle development, significant disparities exist between Luchuan and Duroc pig breeds. In this study, conducted in September 2021 and published in Frontiers in Nutrition with an Impact Factor of 6.57, the authors focused on sampling the Longissimus muscle from both Luchuan and Duroc pigs during the same developmental period. Subsequently, ATAC-seq and RNA-seq analyses were performed.
The study involved correlating differentially accessible chromatin regions identified in ATAC-seq with differentially expressed genes identified in RNA-seq. Following the identification of overlapping genes, the authors conducted annotation and enrichment analyses. This comprehensive approach led to the identification of key genes associated with skeletal muscle development. To validate these findings, RT-qPCR was employed.
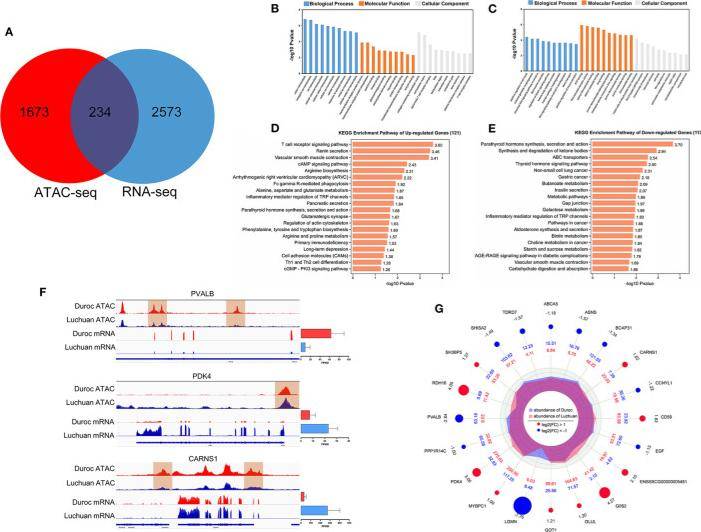 Analysis results of integrated ATAC-seq and RNA-seq results.
Analysis results of integrated ATAC-seq and RNA-seq results.
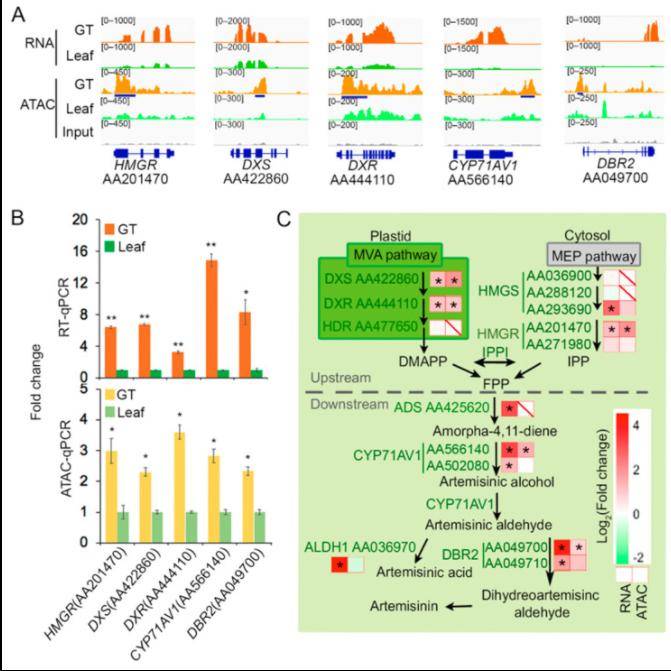
Chromatin Accessibility Is Associated with Artemisinin Biosynthesis Regulation in Artemisia annua
Published: 2021
Journal: Molecules (Impact Factor: 4.4)
To unravel the intricate regulatory mechanisms underlying artemisinin biosynthesis, the author conducted ATAC-seq and RNA-seq analyses on distinct anatomical regions. Their integrated analysis yielded a collection of candidate genes, subsequently subjected to functional and metabolic pathway annotations. Notably, they highlighted the pivotal genes involved in artemisinin synthesis.


 Sample Submission Guidelines
Sample Submission Guidelines
