A Guideline of Genetic Map Construction for Different Populations
What is Genetic Map?
Genetic map construction for different populations is a fundamental process in plant genetics research, allowing us to gain insights into the arrangement and linkage of genes on chromosomes. Also referred to as a genetic linkage map, it is built by analyzing the connections between genetic markers within a population.
The basis of genetic map construction lies in the concept of genetic linkage. When two loci, usually represented by genetic markers, are situated closer together on a chromosome, they tend to be inherited together more frequently. By studying the genetic markers, such as molecular markers like microsatellite markers and SNP markers, or biological markers like morphological and biochemical traits, across various individuals, we can statistically deduce the degree of linkage between them.
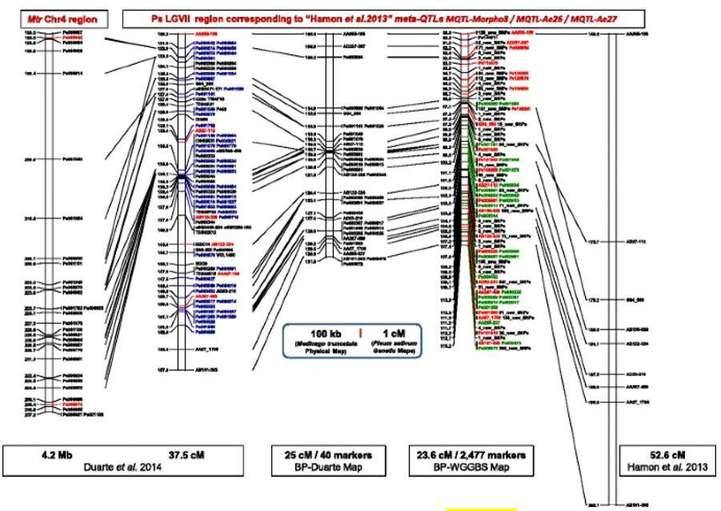 Mapping a RIL population segregating for partial resistance to A. euteiches. (Boutet et al., 2016)
Mapping a RIL population segregating for partial resistance to A. euteiches. (Boutet et al., 2016)
Researchers analyze a significant number of genetic markers and linkage frequencies between parental individuals to draw genetic maps. These maps can take the form of linkage group maps or physical maps, both representing the relative positions of loci on the chromosomes. The distances between loci are usually measured in genetic units known as centimorgans or map units. This information helps determine the sequence and localization of genes on the chromosomes.
The applications of plant genetic maps are vast and vital in genome research, variety improvement, and gene localization. They serve as indispensable tools for understanding the genetic structure and properties of plant genomes, providing valuable insights for researchers seeking to comprehend the genetic basis of specific plant traits. Furthermore, genetic maps offer invaluable guidance for molecular breeding initiatives, contributing to the development of improved and desirable plant varieties.
Please read our article Genetic Linkage Mapping: Definition, Techniques, and Applications for more details.
Genetic Linkage Mapping in Plant Research
Plant genetic mapping plays a pivotal role in elucidating essential aspects of plant genetics across various domains:
- Genetic linkage: This methodology entails the determination of the relative positions of genes on chromosomes by analyzing a plethora of genetic markers, encompassing molecular or genetic markers. Consequently, genetic mapping constructs comprehensive genetic maps, which unveil intricate interrelationships between genes.
- Gene localization: Through genetic mapping, the precise location of a gene of interest on a chromosome can be ascertained. By meticulously analyzing genotypic and phenotypic data, specific genes can be precisely pinpointed and associated with particular traits of interest.
- Gene targeting: Genetic mapping enables the discernment of the direction of gene transfer, involving the identification of sources and targets of gene transmission. This valuable knowledge significantly contributes to the comprehension of gene flow, relatedness, and the intricate patterns of gene transmission within a given population.
- Kinship and lineage analysis: Leveraging genetic mapping, comprehensive analyses of kinship and lineage between individuals, such as father-son, brother-brother, and ancestor relationships, can be conducted. This information holds paramount importance in the effective management of germplasm resources, the formulation of proficient breeding strategies, and making informed parental selections.
Mutation and polymorphism: Genetic mapping proves instrumental in unearthing mutations and polymorphisms prevalent within plant populations. Through a comprehensive assessment of genetic variation at various loci, it provides invaluable insights into the genetic diversity and variation existing within the population. This knowledge, in turn, greatly aids in the conservation of germplasm resources and the advancement of genetic improvement efforts.
Mapping Populations
Various groups are utilized in genetic mapping, with common mapping populations including F1 mapping populations, F2 populations, RIL (recombinant inbred line) populations, and DH (doubled haploid) populations. F1 mapping populations are among the most widely used groups for constructing genetic linkage maps. These populations are formed by crossing two parents, resulting in the first generation of progeny. In the F1 generation, individuals typically exhibit dominant traits, making them easier to observe and measure. Since the F1 generation inherits an equal number of genes from both parents, it is valuable for analyzing genetic mapping and determining linkage between marker loci. The F1 generation's simplified genotypes reduce analysis complexity and increase the stability of genetic mapping.
The F2 Mapping Population
The F2 mapping population represents the second generation of progeny resulting from selfing or crossbreeding (cross-hybridization) of F1 individuals. F2 individuals are produced by selfing or crossbreeding F1 individuals, whereas F1 individuals are obtained by crossing two parents. Self-crossing involves mating individuals within the same F1 generation, while intercrossing refers to mating individuals from different F1 generations. The F2 generation individuals have more complex genotypes with increased genetic variation since they inherit an equal number of genes from their four grandparents. These traits lead to the following characteristics of the F2 generation mapping population:
- Expression of recessive traits: Due to the increased complexity of genotypes in the F2 generation, recessive traits may manifest and require careful observation and measurement.
- Higher genetic diversity: F2 generation individuals exhibit greater genetic variation compared to the F1 generation, facilitating better exploration and analysis of interlocking relationships between loci.
- Increased individual variation in genotypes and phenotypes: The F2 generation mapping population's greater individual variation provides more variability and information for genetic mapping.
RIL Mapping Populations
RIL mapping populations are pure populations obtained by selfing two inbred parents using sequential selfing methods to construct genetic linkage maps. This population type is commonly used in genetic studies and involves the following steps:
- Initial cross: Two different self-fertilizing pure strains or lines are selected as parents and crossed to produce F1 generation progeny.
- Successive selfing: Starting from the F1 generation, successive selfing operations are performed, leading to each generation of individuals being self-pollinated or self-crossed to produce self-fertilized individuals.
- Population establishment: After several generations of selfing, the genotypes among individuals stabilize and become fixed, resulting in a large number of selfed individuals that form a RIL mapping population.
- Marker analysis: Individuals in a RIL population are genotyped using DNA markers or other molecular markers such as SNP markers, SSR markers, and locus-specific sequences.
- Construction of genetic linkage map: Analyzing the genotypes and related phenotypic data of individuals in the RIL population, computational methods and statistical models are employed to infer the linkage relationships and distances between different marker loci, ultimately constructing the genetic linkage map. These maps provide valuable insights into the location of loci, linkage relationships, and segregation, which have significant applications in studying the genetic basis of complex traits, gene localization, and correlation analysis.
DH Mapping Populations
Genetics researchers commonly utilize DH (Doubled Haploid) mapping populations to construct genetic linkage maps. These populations enable asexual reproduction of cells and chromosome purification, leading to the development of self-compatible plants or animals.
The process of creating and employing DH mapping populations involves the following steps:
- Initial hybridization: Two parent individuals, typically self-pure individuals with high affinity, are chosen for hybridization.
- Asexual reproduction: The hybrid individuals undergo appropriate asexual reproduction techniques like pollen culture, embryo culture, haploid induction, and in vitro culture. These methods yield asexually reproduced individuals containing genetic material from only one parent, possessing a haploid number of gamete chromosomes in their genome.
- Chromosome purification: Various chromosome purification techniques are applied to double the number of chromosomes in a haploid individual, transforming it into a doubled haploid individual. Common methods include chromosome doubling, restoration of male fertility from male sterility, and pollen treatment. This process results in a group of self-crossed and pure haploid individuals, forming the DH mapping population.
- Marker analysis: Molecular marker techniques, such as SNP (single nucleotide polymorphism) markers, SSR (simple sequence repeat) markers, etc., are employed for genotype analysis of individuals in the DH population. Genotype data is obtained through gene chips, PCR amplification, and other means.
- Construction of genetic linkage map: Through computational methods and statistical models, researchers analyze the genotypes and related phenotypic data of individuals in the DH population. This analysis helps infer the linkage relationships and distances between different marker loci, allowing the construction of a genetic linkage map.
Utilizing DH mapping populations significantly shortens the time required for self-crossing and purification, leading to a highly pure population. As a result, DH mapping populations have become a powerful tool for rapidly constructing genetic linkage maps and facilitating quick gene localization.
Reference:
- Boutet, Gilles, et al. "SNP discovery and genetic mapping using genotyping by sequencing of whole genome genomic DNA from a pea RIL population." BMC genomics 17.1 (2016): 1-14.


 Sample Submission Guidelines
Sample Submission Guidelines
