Genetic Linkage Mapping: Definition, Techniques, and Applications
Introduction to Genetic Linkage and Linkage Map
DNA sequences that are close together on a chromosome are inclined to be passed down together during the meiosis phase of sexual reproduction. When two genetic markers are physically close to each other, they are less likely to be separated onto various chromatids during the chromosomal crossover and are thus said to be more linked than markers that are far apart. In other words, the closer two genes are on a chromosome, the less likely they are to recombine, and the more likely they will be inherited together. Different chromosome markers are completely unrelated.
A genetic linkage map describes the position of a species' or experimental population's known genes or genetic markers in terms of recombination frequency rather than a particular physical distance along each chromosome. A linkage map is a graph that shows how frequently markers recombine during homologous chromosome crossover. The more recombination (segregation) occurs between two genetic markers, the further apart they are thought to be. Conversely, the lesser the physical distance between the markers, the lesser the frequency of recombination between them. Previously, distinguishable phenotypes (enzyme production, eye color) derived from coding DNA sequences were used as markers; eventually, noncoding DNA sequences such as microsatellites or those generating restriction fragment length polymorphisms (RFLPs) were utilized.
Researchers can use linkage maps to find other markers, such as genes, by testing for genetic linkage between previously identified markers. The data are used to assemble linkage groups, which are groups of genes that are known to be linked, in the early stages of developing a linkage map. More markers can be incorporated into a group as knowledge advances until the group encompasses a whole chromosome.
Recombination frequency
The recombination frequency serves as a metric for genetic linkage, employed in the creation of genetic linkage maps. Genes that are not completely linked invariably undergo recombination due to the exchange of homologous chromosomal segments during meiosis. This leads to the generation of recombinants or progeny characterized by a new combination of traits. These recombinants constitute a certain proportion of the total progeny, represented as a recombination value, percentage of recombination, or recombination frequency.
This measure, expressed as the percentage of recombinants in the total progeny, describes the likelihood of crossover events between given linked genetic markers. Such a benchmark can also be termed as a centimorgan (cM), the unit that illustrates a 1% recombination frequency.
This recombination information provides an indirect, yet powerful, means to quantify the genetic distance between two loci. By examining their recombination frequency, scientists can make inferences about their relative positions on the chromosome, contributing to our understanding of genome architectures and evolutionary dynamics.
Factors Influence the Recombination Frequency: Gene Distance and Position, Transmission Rate and Viability of Gametes, Chromosomal Structural Variations, Hormonal Levels, Temperature and Radiation, Ion Concentration, External factors such as age and etc.
Key points:
- Genes located either on different chromosomes or at considerable distances on the same chromosome, assort independently from each other and are considered non-linked.
- When genes are in close proximity on the same chromosome, they are said to be linked, indicating that alleles or gene variants already present on a chromosome are more likely to be inherited together as a unit.
- By utilizing the data obtained from genetic crossing over to calculate recombination frequencies, we can gain insights into whether two genes are related and ascertain the intensity of their linkage.
- By examining recombination frequencies of numerous pairs of genes, we can construct a linkage map that reveals the order and relative distances of genes on a chromosome.
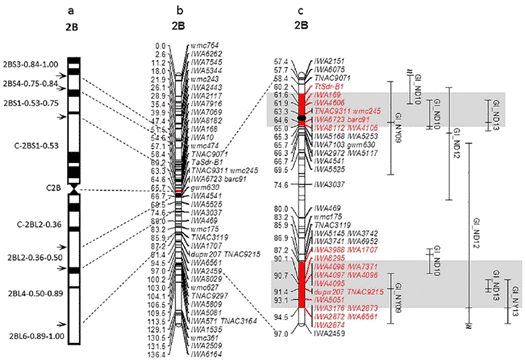 Figure 1. (a) Physical and (b, c) genetic linkage maps. (Chao, 2015)
Figure 1. (a) Physical and (b, c) genetic linkage maps. (Chao, 2015)
Difference Between Genetic Map and Physical Map
A genetic map of a species portrays the relative positions of known genes and/or genetic markers on the chromosome, not their unique physical locations on each chromosome. Using genetic analysis methods such as hybridisation experiments and familial lineage analysis, genes or other DNA markers are ordered on the chromosome. The distance between markers, or the genetic map distance, is represented by the frequency of crossover events during meiosis, this is measured in centimorgans (cM), with each unit defining a 1% exchange rate. The resolution of a genetic map is somewhat coarse, approximately reaching a level of one million base pairs (1Mb).
A physical map, in essence, shows the physical location of identifiable markers (such as restriction enzyme cutting sites or genes) on the DNA. The map distance is the physical length unit, such as the chromosome band or the number of nucleotide pairs.
Comparison of the two:
- A genetic map is based on recombination frequencies, while a physical map is based on directly measured DNA structural features.
- The frequency of meiotic recombination is uneven along the majority of chromosomes, with areas of hotspot and coldspot recombination and/or mutation.
- A genetic map indicates the relative distance between genes or markers, represented by the recombination value and measured in cM.
- A physical map highlights the physical distance between genes or markers, the distance unit is a length unit, such as micrometers (μm) or base pair numbers (bp or kp).
Techniques Used in Genetic Linkage Mapping
Genetic linkage mapping technology primarily encompasses two facets: the detection and analysis of genetic markers, followed by the construction and delineation of linkage maps. In terms of genetic marker detection, modern biological techniques such as Polymerase Chain Reaction (PCR) and sequencing technologies offer high-throughput, high-resolution approaches; these can rapidly and precisely identify a multitude of genetic markers. Conversely, when building linkage correlations, prevalent techniques include genetic linkage analysis, and genotype-phenotype correlation analysis.
Technologies Based on Molecular Markers:
Molecular markers can be categorized into four types according to their technical characteristics: DNA marker techniques based on molecular hybridization, such as Restriction Fragment Length Polymorphisms (RFLP); DNA marker techniques based on PCR, such as Simple Sequence Repeat (SSR); DNA marker techniques combining PCR with restriction enzyme cutting, such as Amplified Fragment Length Polymorphism (AFLP); and DNA marker techniques anchored on Single Nucleotide Polymorphism (SNP).
RFLP: RFLP identifies DNA polymorphisms by cleaving DNA with restriction endonucleases and subsequently detecting varied-length DNA fragments through methods like gel electrophoresis.
AFLP: AFLP integrates the use of restriction endonucleases and PCR to efficiently detect polymorphic fragments within DNA and is often utilized in genetic linkage analysis and genetic mapping.
SSR or Microsatellite Markers: SSR comprises repeated units of short-sequence DNA. Its length variation is handy for genetic linkage analysis. Microsatellite markers, on the other hand, bank on PCR amplification and gel electrophoresis techniques to detect polymorphisms in SSR regions.
SNPs: SNPs can be detected by PCR, sequencing, etc., and are widely applied in genetic linkage analysis and genome-wide association studies.
Technologies Leveraging High-throughput Sequencing:
Genotyping-by-sequencing (GBS): GBS utilizes high-throughput sequencing technology to sequence genomes and perform genetic linkage analysis by observing DNA sequence variations in each individual. This technique is suitable for broad-scale genotype analysis and genetic map establishment.
Whole Genome Sequencing (WGS) for Linkage Mapping: By sequencing the entire genome, WGS can accurately pinpoint the chromosomal locations of genetic markers, providing a high-resolution approach to genetic linkage mapping.
Advanced Computational Methods for Linkage Mapping Analyses:
Genetic Mapping Software and Algorithms: This primarily includes software like JoinMap, MapMaker and algorithms including LOD analysis and Maximum Likelihood methods.
Integration of Linkage Mapping with Genome-wide Association Studies: Combining linkage mapping with GWAS can more precisely discern relationships between genes and phenotypes, and identify new genetic variants relating to specific phenotypes.
Applications of Genetic Linkage Mapping
Positioning and Identification of Genes and Quantitative Trait Loci (QTL): By analysing the genetic linkage relationships, the chromosomal locations of genes influencing specific traits can be determined, enabling their positioning and identification. This uncovers essential clues and tools for understanding the genetic basis of various traits within organisms. Among the classical research methodologies is QTL mapping, a process that identifies the genes related to the target traits based on the linkage relationship between molecular markers and the target traits. Genetic mapping is typically employed for locational analyses of shape variations between two parents with markedly different traits and offspring populations with multiple phenotypic traits.
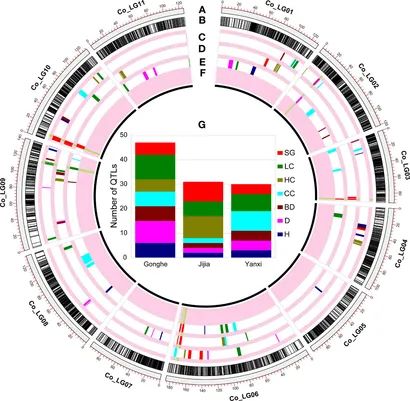 Figure 2. QTL mapping in multiple environments. (Fang et al., 2020)
Figure 2. QTL mapping in multiple environments. (Fang et al., 2020)
Marker-assisted Breeding: Through the analysis of genetic markers related to the target traits, breeders can more precisely choose individuals with the desired traits for breeding, thus enhancing the efficiency and speed of selective breeding. This technique enables breeders to predict and select individuals with desirable traits without extensive laboratory or field trials, presenting an effective strategy for crop quality and yield improvement. Based on the construction of a genetic linkage map, further gene positioning can be carried out through marker semination and phenotype identification, thereby obtaining the target gene. This establishes a solid foundation for further gene cloning and its applications in breeding.
The genetic mapping of plants is widely applied in genomics research, variety improvement, and gene positioning. It offers an important tool for understanding the structure and genetic properties of plant genomes, providing valuable guidance for researchers to delve into the genetic basis of plant traits and carry out molecular breeding. Bernard et al. employed techniques such as genetic linkage mapping to investigate the genetic structure of phenological traits and lateral bearing in Persian walnut (Juglans regia L.). Their findings enhanced our understanding of the genetic architecture underlying important agronomic traits associated with walnut male and female flowering processes as well as lateral bearing. The results underscored the significance of germination date as one of the most crucial traits for desirable outcomes, suggesting that novel markers would aid in the selection and development of walnut varieties suited to specific climates.
Comparative Mapping and Evolutionary Studies: By comparing the genetic linkage maps of different species or populations, their genetic relationships, evolutionary histories, and population structures can be revealed. This aids researchers in understanding the origin, evolution, and adaptive evolutionary processes of species, offering important clues in the protection of biological diversity and the exploration of evolutionary biology mechanisms.
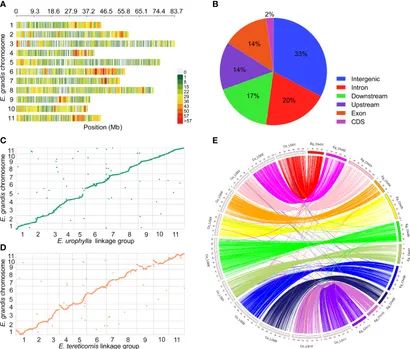 Figure 3. SNP calling and genetic mapping construction. (Fang et al., 2020)
Figure 3. SNP calling and genetic mapping construction. (Fang et al., 2020)
Identification of Genetic Variants Associated with Disease in Humans and Model Organisms: By analysing genetic linkage maps, the chromosomal position of genetic variants associated with diseases can be determined, and their influence on disease onset and development can be studied further. This provides an important foundation and basis for early disease diagnosis, risk assessment, and treatment, bearing significant implications for human health and disease prevention and control. Julier et al. conducted an investigation into the genetic map of human chromosome 22, elucidating the localization of the P1 polymorphism of the P blood group to chromosome 22 (proximal to the SIS oncogene locus), thereby furnishing additional evidence.
References:
- Chao S, Elias E, Benscher D, et al. Genetic mapping of major-effect seed dormancy quantitative trait loci on chromosome 2B using recombinant substitution lines in tetraploid wheat. Crop Science. 2016 Jan;56(1).
- Fay D. Genetic mapping and manipulation: Chapter 1-Introduction and basics. WormBook. 2006 Feb 17;17.
- Jeffreys A J, Neumann R. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Human molecular genetics, 2005, 14(15): 2277-2287.
- Julier C, Lathrop G M, Reghis A, et al. A linkage and physical map of chromosome 22, and some applications to gene mapping. American journal of human genetics, 1988, 42(2): 297.
- Bernard A, Marrano A, Donkpegan A, et al. Association and linkage mapping to unravel genetic architecture of phenological traits and lateral bearing in Persian walnut (Juglans regia L.). BMC genomics, 2020, 21: 1-25.
- Fang Y, Zhang X, Zhang X, et al. A high-density genetic linkage map of SLAFs and QTL analysis of grain size and weight in barley (Hordeum vulgare L.). Frontiers in Plant Science, 2020, 11: 620922.


 Sample Submission Guidelines
Sample Submission Guidelines
