Genome Assembly and Annotation: Background, Workflow and Applications
Genome assembly and annotation are essential components in comprehending the genetic blueprint of life. Genome assembly is the reconstruction of an organism's full DNA sequence into a continuous and coherent strand, while annotation is the process of assigning functional roles to these sequences by identifying genes, regulatory elements, and other important features. These sequences then undergo various processes to derive meaning and insights from the data, illuminating patterns in human evolution, medicine, and biotechnology.
What is Genome Assembly and Annotation
Emerging sequencing technologies have rapidly simplified the pipelines for genome assembly and annotation, empowering scientists to tackle genomes of unprecedented complexity. Extensive use of new computational tools and algorithms enabled scientists to assemble even difficult genomes, such as those with high repeat content or polyploid. This progress has established genome assembly and annotation as essential means of investigating a range of genetic architectures, from microbial communities and human genomes to plant breeding projects. In this article, we explore the tenets, methods, and uses of these interrelated processes, which offer tremendous promise for the discipline of genomics.
Background in Genome Assembly and Annotation
Genome Assembly and Genome annotation
For some background, genome assembly is the process of piecing together an organism's genome DNA into its complete sequence from short DNA sequences, called reads. This involves aligning millions to billions of sequencing reads into continuous sequences (contigs), and ordering contigs into scaffolds representing chromosomes. The treatment is not straightforward computational task, and require advanced algorithm to handle sequencing errors, repeative region and genomics variation.
Genome annotation is the process of identifying and labeling the functional elements of a genome. It typically includes sequencing the genomes, predicting genes and non-coding elements, and studying regulatory elements, non-coding RNAs and repetitive elements. With annotation, an otherwise inanimate sequence transforms into an on-demand annotation resource for biologists interested in obtaining and summarizing functional information regarding genes, regulatory networks, and evolutionary relationships.
Service you may interested in
Resource
Types of Annotation
Structural Annotation: Identifies genes, exons, introns, regulatory sequences and repeat elements. Prediction of coding regions and annotation of genomic elements.
Functional Annotation: Biological role assignment for identified features by similarity to known genes, protein domains and pathways. Functional annotation provides added insight into gene functionality and biological systems.
Annotation Tools and Techniques
Structural Annotation Tools: AUGUSTUS, GeneMark, etc. These tools utilize probabilistic models to detect genes from alignments of genomic sequences.
Functional Annotation Tools: Avoid filtering critical loci, databases such as UniProt and GO (Gene Ontology) provide functional insights, while tools like BLAST align sequences to known genes for comparative annotation. Functional annotation is further enhanced by a linkage to metabolic and regulatory networks via pathway databases such as KEGG.
Automated Pipelines: Tools such as MAKER and Prokka are designed to combine various prediction and alignment tools to provide a comprehensive annotation workflow. Such automated pipelines have substantially decreased the annotation time of large genomes allowing researchers to concentrate on downstream analysis and applications.
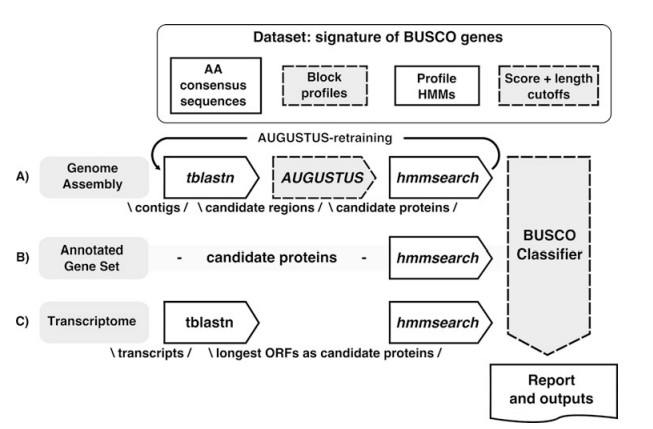 Description of the BUSCO workflow (Seppey, M. et al 2019).
Description of the BUSCO workflow (Seppey, M. et al 2019).
Genome Assembly and Annotation Workflow
Genome Assembly Pipeline
Data preprocessing: Quality control, trimming and error correction are performed to ensure that only high-quality reads are used for assembly. FastQC, Trimmomatic, and Racon are some of the other tools commonly used to clean and polish sequencing data and correct common errors such as sequencing errors and adapter contaminants.
Assembly: Contigs and scaffolds are assembled from reads, using de novo, reference-guided, or hybrid approaches. SPAdes, Flye and Canu are examples of assemblers that use sophisticated algorithms to yield more accurate and contiguous sequences. Contigs are ordered and oriented into scaffolds, and remaining errors are corrected to enhance base-level accuracy. These software tools, such as LINKS, SSPACE, and Pilon, fill in gaps in the sequence and improve assembly quality.
All approaches suggest that assembly quality is assessed using metrics like N50, BUSCO completeness scores, and alignment accuracy. These statistics give information about the quality and completeness of the genome assembly.
Genome Annotation Pipeline
Repeat Masking: It mask out all the repetitive sequences which may give false-positive predictions. Repetitive elements can be annotated and managed using well-known tools such as RepeatMasker and Tandem Repeats Finder.
Gene Prediction: Tools that predict coding and non-coding genes from sequence patterns, homology or statistical models. Ab initio tools such as AUGUSTUS predict genes from sequence features (such as coding and non-coding sequence) whereas evidence-based tools use transcriptomic or proteomic data.
Functional Annotation: We aligned the predicted genes to known databases to assign functions to features. Functional annotation tools help to understand genes and their roles, giving context to genomic data in relation to biological pathways and systems.
High-Value Manual Review: Hand curation by quality reviewers for any regions or genes with high importance This is especially important for genes of medical or agricultural interest.
Genome Assembly & Annotation Applications
Medicine
Genomic assembly and annotation are crucial to locating disease-related genes, deciphering disease mechanisms, and devising personalized therapies. For instance, genome sequencing and annotation of pathogenic microbes supports rapid diagnostics and vaccine development. The annotated genomes serve as a foundation for identification of gene-disease associations, diagnostic biomarkers, and therapeutic targets.
Agriculture
Characterized crop genomes have stimulated precision breeding by mapping genes associated with yield, disease resistance, and stress tolerance. High-confidence genome assembly and annotation of the wheat genome have revealed important genes for traits such as drought tolerance and pest resistance. Such insights have revolutionised farming methods, allowing scientists to breed resilient varieties of crops.
Evolution and Ecology
Genome assembly and annotation allow comparative analyses that enable to uncover evolutionary relationships, speciation events and adaptive traits. Annotated genomes of endangered species inform conservation strategies by assigning attributes to specific genes that promote genetic diversity and resilience. The study of adaptive evolution and understanding the genetic bases of adaptation allow researchers to create more specific conservation and management approaches.
Case Study: Annotating the Arabidopsis Genome
Background
Arabidopsis thaliana is one of the most widely used model organisms in plant biology and one of the first plant genomes to be sequenced and annotated. Its relatively small genome (∼135 Mb) and simple structure made it a model target for early genome projects. The assembly and annotation of Arabidopsis established the basis for plant genomics and propelled advances in both functional genomics and crop improvement.
Methods
Sequencing: The genome was sequenced with Sanger technology, and was then refined with high-throughput. A combination of sequencing strategies was used to maximize coverage and accuracy.
Assembly: An initial genome was built using de novo assembly and then improved through multiple polishing rounds. Plant genomes are framed through tools customized for them to resolve repeat regions and structural complexities.
Annotation: Structural annotation yielded ~27,000 protein-coding genes Functional annotation Most of the genes were assigned to a function through homology-based methods, integrating data from several functional databases.
Results
The annotated Arabidopsis genome has had an immense impact on plant genomics, informing research in gene function, regulatory networks, and plant-environment interactions. Its high-quality annotation has also enabled comparative studies with crop species that promote agricultural innovation. The detailed annotation of Arabidopsis embodies a reference point for the unique exploration of basic biological questions in plant science.
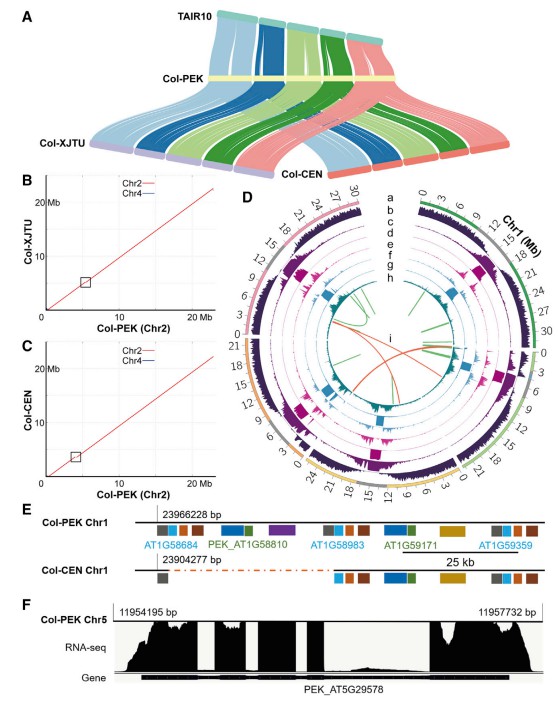 Overview of Col-PEK assembly (Hou, X. et al, 2022).
Overview of Col-PEK assembly (Hou, X. et al, 2022).
Methodological Advancements in Genome Assembly and Annotation
Novel Sequencing Platforms
Ultra-long reads and single-cell sequencing are allowing chromosome-level assemblies and high-resolution annotation of complex genomes. These developments can potentially solve genomes regions that were inaccessible previously, like centromeres and telomeres. Combining new sequencing technologies enable researchers to break through traditional barriers to genome assembly and annotation.
Integration with Multi-Omics
Integrated approaches that combine genome annotation with transcriptomics, proteomics, and epigenomics can lead to a more holistic view of gene function and regulation. This integrative strategy is especially informative for the investigation of dynamic biological processes and complex traits. By connecting the sequence to the phenotype, multi-omics data integration allows the functional interpretation of genomic data.
Service you may interested in
AI and Machine Learning
These advances in artificial intelligence are revolutionizing annotation how by guiding gene prediction, functional assignment, and error correction. AI-powered tools can also process enormous datasets at an unparalleled scale and speed — a capability that can significantly accelerate genome annotation efficiency. Multitude of genomic datasets have been used to train machine learning models that can serve as predictive tools facilitating annotations of currently sequenced genomes.
Global Genomics Initiatives
Initiatives such as the Earth BioGenome Project are aiming towards sequencing and phenotyping the genomes of all eukaryotic species. These efforts are promoting collaboration, standardizing workflows, and democratizing access to genomic data.” The emergence of shared genomic databases is speeding up discovery and enabling the exploration of biodiversity and ecosystem functions.
Conclusion
Genome assembly and annotation is one of the most important areas for many modern genomics applications, as it enables researchers to make sense of the functional potential within the genome from DNA sequences. Facilitated by the combination of new sequencing methodologies with high-performance computing applications, these processes have drastically improved our ability to understand genetic structure and its implications from a biological, medical, and agricultural standpoint. Genome assembly and annotation are critical steps that underpin genomic research and will continue to evolve in the field, enabling scientific discovery to remain central to our progress in biology and in addressing global problems. Global initiatives and emerging technologies are likely soon to expand the range and utility of these critical genomic resources.
References:
- Seppey, M., Manni, M., & Zdobnov, E. M. (2019). BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods in molecular biology (Clifton, N.J.), 1962, 227–245. https://doi.org/10.1007/978-1-4939-9173-0_14
- Hou, X., Wang, D., Cheng, Z et al. (2022). A near-complete assembly of an Arabidopsis thaliana genome. Molecular plant, 15(8), 1247–1250. https://doi.org/10.1016/j.molp.2022.05.014


 Sample Submission Guidelines
Sample Submission Guidelines
