What is Gene and Gene Sequencing?
What is a Gene?
A gene serves as a fundamental unit of genetic information encoded within DNA. It orchestrates the expression of an organism's traits by guiding protein synthesis through the intricate processes of transcription and translation. In essence, genes dictate the manifestation of an individual's genetic makeup, wielding profound influence over the traits exhibited by an organism.
What is Gene Sequencing?
Gene sequencing unveils the intricate blueprint of life encoded within our genes. Every organism's genetic makeup, while diverse, ultimately reflects a unique arrangement of the four fundamental bases: adenine (A), thymine (T), guanine (G), and cytosine (C). At its essence, gene sequencing is the meticulous process of unraveling this sequence, illuminating the precise order of these bases within a gene.
The technique harnesses the power of a gene sequencer, a sophisticated instrument designed to decode the genetic information embedded within DNA. Employing a nuanced approach, gene sequencing utilizes fluorescent markers of distinct colors to distinguish between the four bases. As the gene passes through the sequencer, a laser system triggers the emission of fluorescent light from each base, revealing their sequential arrangement.
These emitted lights, each corresponding to a specific base, are then captured by a specialized camera, transforming the genetic sequence into a visual tapestry of vibrant hues. Through meticulous analysis of these fluorescent signals, the precise order of bases within the gene is deciphered, offering profound insights into the fundamental building blocks of life.
CD Genomics high-throughput sequencing and long-read sequencing platforms facilitate the robust analysis of DNA/genomes. This advanced sequencing approach allows for comprehensive and efficient examination of genetic material, providing valuable insights into the molecular landscape and potential biomarkers associated with various conditions.
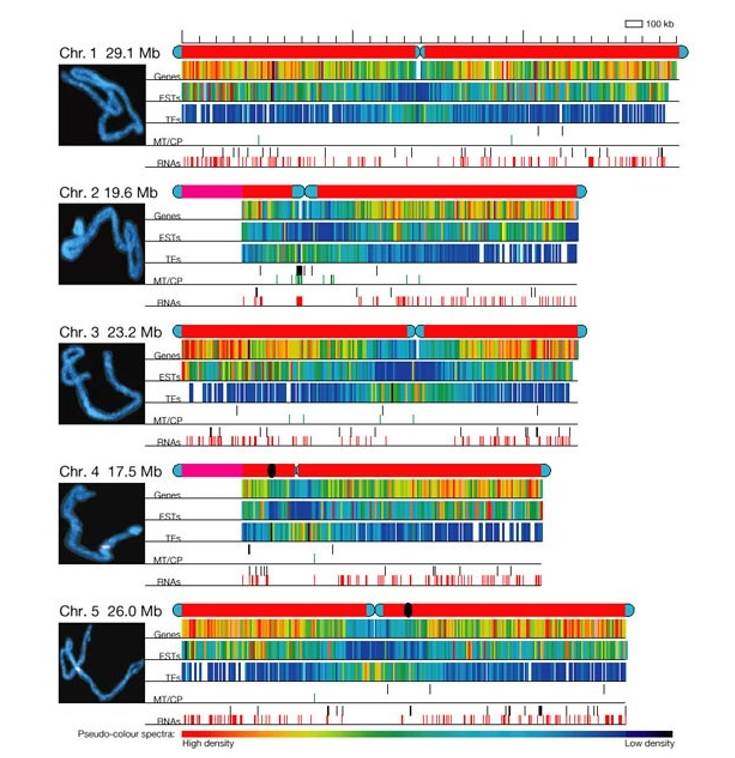 Representation of the Arabidopsis chromosomes. (Iniative et al., 2000)
Representation of the Arabidopsis chromosomes. (Iniative et al., 2000)
Applications of Gene Sequencing
Gene sequencing has become ubiquitous, revolutionizing our approach to understanding and managing genetic information. This cutting-edge technology analyzes and deciphers the complete sequence of genes extracted from blood or saliva samples, offering invaluable insights into disease predisposition, behavioral traits, and individualized medical care.
With the ability to predict the likelihood of various diseases and discern behavioral characteristics, gene sequencing plays a pivotal role in preemptive healthcare. By identifying genetic markers associated with specific conditions, it enables proactive measures to prevent and treat diseases before they manifest.
Over 45 years, gene sequencing has undergone remarkable evolution, culminating in the fourth generation of sequencing technology. Advancements in sequencing techniques, coupled with declining costs, have democratized access to this transformative tool, integrating it into everyday life.
The applications of gene sequencing span a diverse array of fields, encompassing both research and clinical domains. From multi-omics studies and population cohort sequencing to drug development, agriculture, and public health management, its impact is far-reaching. In clinical medicine, gene sequencing facilitates non-invasive prenatal testing, assists in reproductive technologies, aids in tumor diagnosis and precision treatment, and enables early detection of infectious diseases.
Today, gene sequencing transcends traditional medical boundaries, finding utility in scientific research, consumer services, and beyond. Its versatility underscores its pivotal role in shaping the future of healthcare and beyond.
The History of Gene Sequencing
The history of gene sequencing spans 45 years, marked by four generations of technological innovation that have propelled the industry's exponential growth.
In 1977, Sanger pioneered the first generation of gene sequencing, laying the foundation for subsequent advancements. A pivotal moment arrived in 1998 with the advent of capillary sequencing technology, which accelerated sequencing speeds tenfold and catalyzed the Human Genome Project's ambitious timeline.
The turn of the millennium ushered in the second generation of high-throughput sequencers in 2005, slashing costs by a staggering 100-fold and igniting a trend that surpassed even Moore's Law, aptly dubbed "super Moore's Law." This era witnessed unprecedented improvements in sequencing speed, length, and throughput.
Today, next-generation sequencing (NGS) stands as the pinnacle of commercial viability and widespread adoption. NGS revolutionized sequencing by introducing reversible termination of unbound nucleotides, enabling simultaneous synthesis and sequencing with unparalleled throughput, precision, and affordability. While innovations like PacBio SMRT sequencing and nanopore technologies promise enhanced accuracy and versatility, the practical challenges of cost and precision persist, ensuring the dominance of NGS in the foreseeable future.
Yet, the trajectory of gene sequencing continues to evolve, driven by the quest for higher throughput, longer read lengths, greater accuracy, faster turnaround times, enhanced fault tolerance, broader applicability, and reduced costs. As technology progresses, the landscape of gene sequencing holds boundless potential for transformative advancements that will shape the future of healthcare and scientific discovery.
Sanger Sequencing Technology
Sanger sequencing, hailed as the gold standard for mutation detection and validation, emerged in 1977 as the pioneering force in genetic analysis.
This Sanger sequencing method employs electrophoresis to separate DNA fragments of varying lengths, enabling precise sequencing. It finds its niche in scenarios requiring meticulous genetic diagnosis, such as familial lines with clearly defined causative genes, small sample cohorts, and known genetic loci.
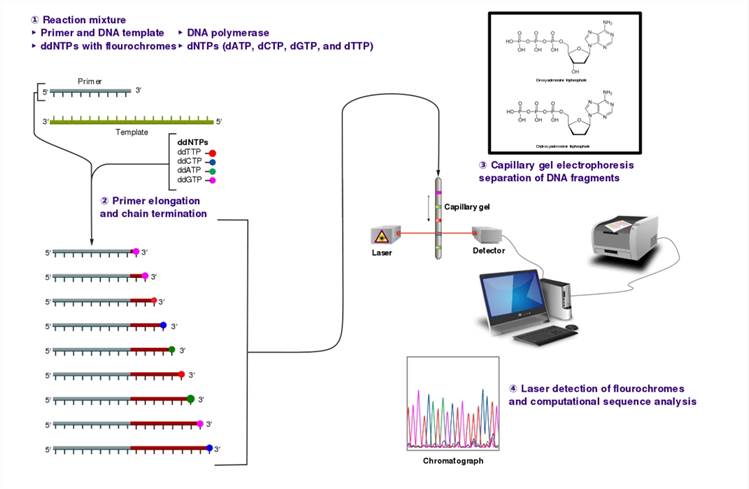 The Sanger (chain-termination) method for DNA sequencing
The Sanger (chain-termination) method for DNA sequencing
The strengths of Sanger sequencing lie in its ability to generate lengthy reads, extending up to 1000 base pairs, coupled with unparalleled accuracy, setting the benchmark for mutation detection and validation. Leveraging PCR amplification, a routine laboratory procedure, enhances its practicality. However, limitations loom, including low throughput, prohibitive sequencing costs, intricate operation procedures, and limited automation, rendering it inadequate for large-scale gene sequencing endeavors.
Moreover, its inability to detect large deletions, disparities in gene copy numbers, and certain mutation types underscores the necessity for reference sequences, further complicating its utility.
CD Genomics Sanger sequencing platform facilitates the robust analysis of DNA/genomes. This advanced sequencing approach allows for comprehensive and efficient examination of genetic material, providing valuable insights into the molecular landscape and potential biomarkers associated with various conditions.
NGS Sequencing Technology
Next-generation sequencing stands at the forefront as the most commercially mature and widely embraced sequencing methodology. Within this realm, five distinct categories emerge: pyrophosphate sequencing, synthetic sequencing, ligation sequencing, semiconductor sequencing, and DNB.
NGS, or next-generation high-throughput sequencing, builds upon the foundation of synthetic termination sequencing, relying on PCR amplification of DNA templates bound to solid surfaces for synthesis and sequencing. This technology finds extensive application in identifying candidate genes associated with diseases, spanning from monogenic disorders to complex conditions like diabetes, obesity, and even cancer.
While next-generation sequencing boasts remarkable throughput, precision, and affordability, its short read lengths pose challenges, potentially leading to the loss of less abundant sequence information during sequencing. Furthermore, except for DNB, reliance on PCR may introduce systematic errors, necessitating repetitive sequencing to mitigate discrepancies.
DNB technology circumvents these issues by minimizing PCR bias, thereby enhancing sequencing accuracy. Consequently, high-throughput sequencing emerges as a cornerstone technology, driving the widespread adoption and commercialization of gene sequencing.
CD Genomics high-throughput sequencing and long-read sequencing platforms facilitate the robust analysis of DNA/genomes. This advanced sequencing approach allows for comprehensive and efficient examination of genetic material, providing valuable insights into the molecular landscape and potential biomarkers associated with various conditions.
Long-Read Sequencing
Single-molecule read-time sequencingrepresents a paradigm shift, utilizing physical methods to decipher genetic information at the individual molecule level. Evolving from NGS, this technology addresses the fundamental limitations of read length and PCR bias inherent in its predecessor.
As a groundbreaking advancement in genome sequencing, PacBio single-molecule read-time sequencing offers unprecedented capabilities for analyzing genetic material at both the single-cell and single-molecule scale.
The long-read sequencing technology extends sequence read lengths beyond those achieved by its predecessors, enabling direct detection of RNA and methylation sequences without the need for PCR amplification. By bypassing PCR, it mitigates the risk of introducing artificial mutations.
However, while PacBio SMRT sequencing excels in many areas, it falls short in single-gene detection applications, particularly for monogenic disorders. Its primary drawback lies in its lower sequencing accuracy, necessitating multiple repetitions or correction via probabilistic algorithms to enhance reliability.
This need for repetitive sequencing or algorithmic correction significantly elevates the cost and diminishes the efficiency of sequencing, posing challenges for widespread adoption and implementation.
Oxford Nanopore Technologies (ONT)has pioneered groundbreaking advancements in sequencing technology, revolutionizing both cost and speed parameters while continually shrinking instrument sizes. Central to ONT's innovation is the nanopore, a minute orifice immobilized on a resistive membrane, with a motorized protein facilitating its movement across a transmembrane electric field.
Within this setup, a DNA single strand is drawn through the nanopore by the motorized protein, propelled from the negative to the positive pole under the influence of the transmembrane electric field. Due to the nanopore's diminutive size, only a single base can traverse it, each base inducing a unique disruption in the electric current, which is subsequently detected and recorded.
Nanopore sequencing boasts several advantages, including extended read lengths, cost-effectiveness, rapid sequencing speeds, and remarkable efficiency. It facilitates dynamic real-time sequencing and supports direct sequencing of native DNA and RNA, achieving detection accuracies ranging from 92% to 98%.
However, despite these strengths, nanopore sequencing falls short of the accuracy achieved by next-generation sequencing methods, with accuracy rates typically ranging from 92% to 98%. Unlike PacBio SMRT sequencing, which employs repeat sequencing to enhance accuracy, nanopore sequencing relies on hydrolysis sequencing methods, contributing to longer sequencing times and constrained accuracy improvement capabilities.
Thus, while Nanopore sequencing offers remarkable advantages, including its versatility and real-time capabilities, its lower sequencing accuracy and limited accuracy enhancement mechanisms pose challenges for fine-tuning sequencing parameters such as time and cost.
CD Genomics high-throughput sequencing and library construction services enable in-depth analysis of transcriptomes. CD Genomics provides short-read and long-read RNA-Seq service, ensuring robust transcriptome research in samples of the highest quality.
Reference:
- Iniative, Arabidopsis Genome. "Analysis of the genome sequence of the flowering plant Arabidopsis thaliana." Nature 408.6814 (2000): 796-815.


 Sample Submission Guidelines
Sample Submission Guidelines
