Next-Generation Sequencing (NGS) Technologies: Advances in Fusion Gene Detection and Targeted Therapy
Fusion Gene
Fusion genes (FGs) resulting from the structural rearrangement of genetic material play a crucial role as oncogenic drivers in various adult and pediatric solid tumors. They have emerged as important molecular markers for tumor diagnosis, treatment, and prognostic assessment.
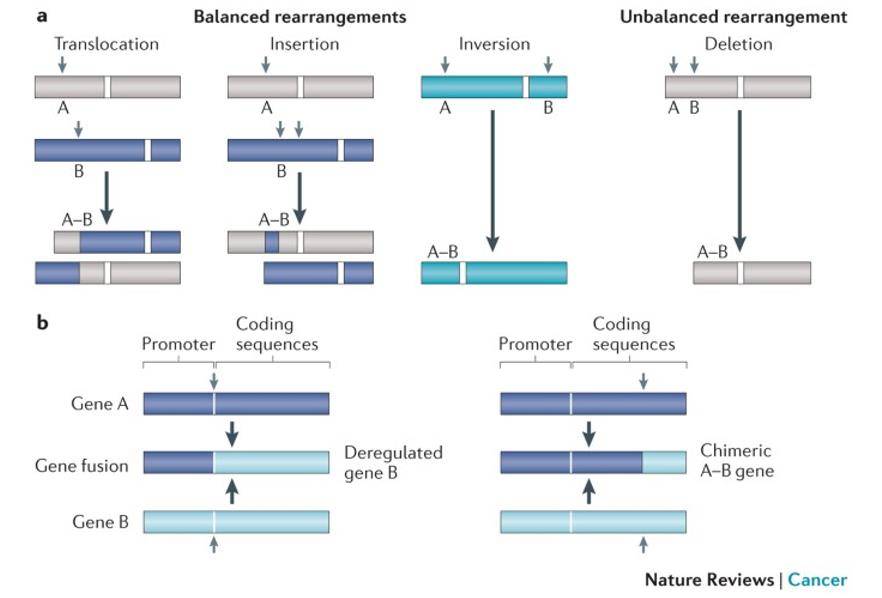 Gene fusion. (Mertens et al., 2015)
Gene fusion. (Mertens et al., 2015)
The detection of fusion genes (FGs) holds significant importance in understanding the molecular landscape of solid tumors. FGs arise from the fusion of genetic material, playing a pivotal role as oncogenic drivers in both adult and pediatric patients. They have emerged as crucial molecular markers for tumor diagnosis, treatment, and prognostic assessment. However, traditional fusion gene detection techniques such as reverse transcription-polymerase chain reaction (RT-PCR) and fluorescence in situ hybridization (FISH) have limitations in detecting unknown breakpoints and complex structural variants.
Next-Generation Sequencing (NGS) and Fusion Gene Detection
NGS technologies have revolutionized fusion gene detection by enabling high-throughput analysis of gene fusions. NGS offers several advantages in fusion gene detection by providing a comprehensive overview of the genetic landscape. However, there are challenges and limitations that need to be addressed. The coverage of intronic regions remains a challenge, as these areas may contain critical breakpoints. Additionally, the sensitivity of NGS to detect low-frequency fusions and the assessment of the weighting impact of differentially expressed RNAs pose further difficulties.
Strategies to overcome these challenges are actively being pursued. Improved library preparation protocols, sequencing depth, and bioinformatics algorithms are being developed to enhance intronic region coverage and detection sensitivity. Integrating additional genomic and transcriptomic data can aid in accurately identifying and interpreting fusion events. These advancements pave the way for comprehensive fusion gene analysis.
Applications of NGS in Fusion Gene Detection
Fusion Gene Network Analysis
NGS data can be utilized to construct fusion gene networks, which provide insights into the functional relationships between fusion genes and other genes within cellular pathways or networks. By integrating gene expression data, protein-protein interaction networks, and functional annotation databases, NGS-based fusion gene network analysis can unravel the complex interplay between fusion genes and downstream signaling pathways, contributing to a deeper understanding of tumor biology.
Long-Read Sequencing for Structural Variation Detection
While short-read NGS platforms are commonly used for fusion gene detection, long-read sequencing technologies, such as Pacific Biosciences (PacBio) and Oxford Nanopore, offer enhanced capabilities in detecting complex structural variations, including fusion genes. Long-read sequencing enables the identification of large genomic rearrangements, novel fusion breakpoints in repetitive regions, and fusion events involving multiple genes or distant genomic loci. This technology provides a more comprehensive view of fusion gene landscapes, especially in cases where conventional short-read sequencing might miss certain structural variations.
Read our article Long-Read Sequencing Data Analysis Service: Advancing Genomic Research with CD Genomics for more information.
Fusion Gene Expression Dynamics
NGS-based techniques, such as RNA sequencing (RNA-seq), can capture the dynamic expression patterns of fusion genes across different stages of tumor development or in response to therapeutic interventions. Time-series RNA-seq data can reveal the temporal dynamics of fusion gene expression, shedding light on their roles in tumor progression, treatment resistance, and disease recurrence. Additionally, single-cell RNA-seq (scRNA-seq) can provide insights into the heterogeneity of fusion gene expression within individual tumor cells, allowing for the identification of rare cell populations and potential therapeutic targets.
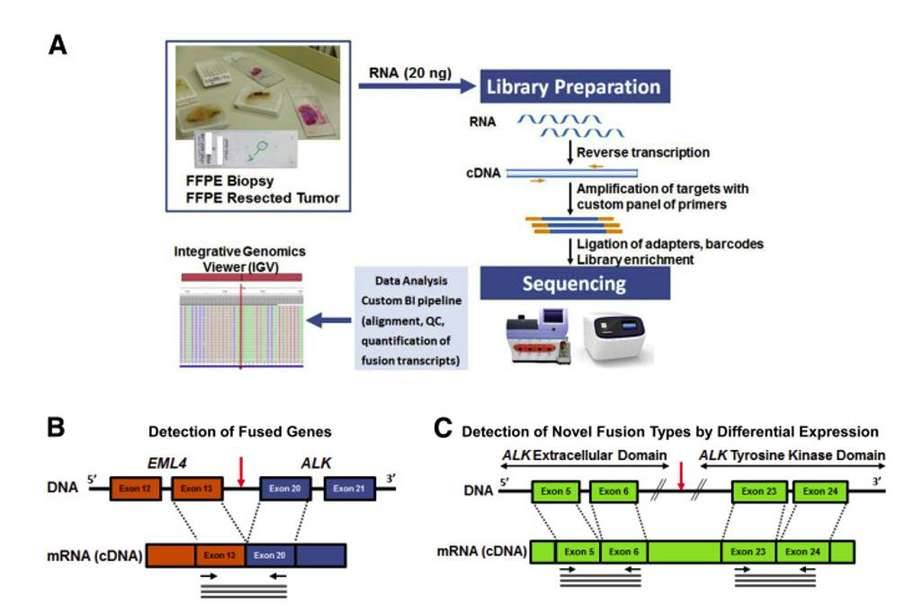 Detecting gene fusions by Next-Generation Sequencing. (Beadling et al., 2016)
Detecting gene fusions by Next-Generation Sequencing. (Beadling et al., 2016)
Integration of Multi-Omics Data
NGS data can be integrated with other high-throughput omics data, such as DNA methylation profiles, chromatin accessibility assays (ATAC-seq), and proteomics data, to unravel the epigenetic and post-transcriptional regulatory mechanisms associated with fusion gene expression. Integrative analysis of multi-omics data can uncover the regulatory elements, transcription factors, and chromatin states that influence fusion gene formation and their functional consequences, leading to a more comprehensive understanding of the underlying molecular mechanisms.
Fusion Gene Validation and Functional Studies
NGS data can guide the design and validation of fusion gene events through targeted approaches such as polymerase chain reaction (PCR), Sanger sequencing, or digital droplet PCR. Functional studies, such as cell line models, organoids, or xenograft models, can be employed to investigate the functional impact of fusion genes on cellular processes, drug sensitivity, and therapeutic resistance. NGS can provide the initial evidence for fusion gene detection, followed by comprehensive validation and functional characterization to elucidate their role in disease biology.
For more information, please refer to Sanger Sequencing: Introduction, Workflow, and Applications.
Targeted Therapies and Fusion Genes
Therapeutic Implications of Fusion Genes in Solid Tumors
Fusion genes are often associated with specific tumor types and can serve as therapeutic targets. Understanding the functional consequences of fusion genes allows for the development of targeted therapies that exploit these specific vulnerabilities in cancer cells.
Overview of Targeted Therapies for Fusion Gene-Driven Cancers
Numerous targeted therapies have been developed to specifically address fusion gene-driven cancers. These therapies include small molecule inhibitors, monoclonal antibodies, and immune-based approaches. They have shown promising results in clinical trials and provide new avenues for personalized treatment strategies.
Challenges and Future Prospects of Fusion Gene-Targeted Therapies
Despite the successes, challenges remain in developing and implementing fusion gene-targeted therapies. Overcoming issues such as acquired resistance and identifying optimal combination therapies are areas of active research. Future prospects involve the integration of multi-omics data and the development of novel therapeutic approaches to improve patient outcomes.
Future Directions and Emerging Technologies
Innovations In NGS-based Fusion Gene Detection Methods
The field of fusion gene detection continues to evolve with ongoing innovations in NGS technologies. Improved library preparation techniques, advancements in sequencing technologies, and novel bioinformatics approaches are being developed to overcome existing limitations. These innovations aim to enhance the sensitivity, accuracy, and efficiency of fusion gene detection, enabling a more comprehensive understanding of tumor biology.
Integration of Multi-Omics Data for Comprehensive Fusion Gene Analysis
The integration of multi-omics data, including genomic, transcriptomic, and epigenomic profiles, holds great promise for comprehensive fusion gene analysis. Integrative approaches can provide a holistic view of fusion gene landscape, unravel complex regulatory networks, and identify potential therapeutic targets beyond gene fusions alone. Such comprehensive analyses have the potential to guide precision medicine strategies and improve patient outcomes.
Potential Impact of Fusion Gene Detection on Precision Medicine
The advancements in fusion gene detection, combined with targeted therapies, have the potential to revolutionize precision medicine. Fusion genes can serve as predictive biomarkers for treatment response and guide the selection of targeted therapies. Furthermore, ongoing research exploring the functional consequences of fusion genes may unveil new therapeutic vulnerabilities and combination treatment strategies. Precision medicine approaches based on fusion gene detection can lead to more tailored and effective therapies for cancer patients.
References:
- Mertens, Fredrik, et al. "The emerging complexity of gene fusions in cancer." Nature Reviews Cancer 15.6 (2015): 371-381.
- Beadling, Carol, et al. "A multiplexed amplicon approach for detecting gene fusions by next-generation sequencing." The Journal of molecular diagnostics 18.2 (2016): 165-175.


 Sample Submission Guidelines
Sample Submission Guidelines
