The DNA methylation analysis technology, EpiTYPER MassARRAY, provided by Agena Bioscience (previously Sequenom Inc.), is one of the most reliable quantitative methods available today for DNA methylation analysis. EpiTYPER is a MALDI-TOF mass spectrometry-based bisulfite sequencing method that enables region-specific DNA methylation analysis in a quantitative and high-throughput fashion. The technology interrogates 10s-100s of samples and CpG sites in amplicons from 200 - 600 bp and detects down to 5% differences in methylation. It is particularly suitable for larger scale efforts to study candidate regions or to validate regions from genome-wide DNA methylation studies.
Workflow - From Assay Design to Results
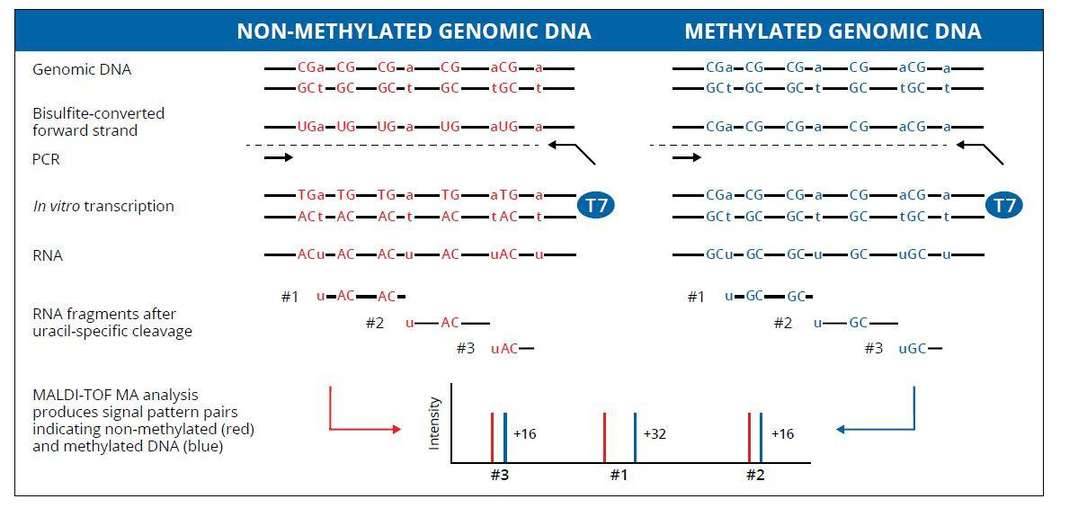
EpiTYPER biochemistry starts with bisulfite treatment of genomic DNA, followed by PCR amplification of target regions. The reverse primers contain a T7 promoter tag. Next, in vitro RNA transcription is performed, followed by base-specific RNA cleavage. Finally, the cleavage products are analyzed using MALDI-TOF mass spectrometry (MassARRAY Analyzer). The methylated and non-methylated cytosine residues in the original genomic DNA are easily distinguished using EpiTYPER Software.
STEP 1: Assay Design
EpiDesigner is an online automated design tool for DNA methylation experiments on the MassARRAY System. Just enter your target sequences and the software determines primer designs for the most complete DNA coverage. In addition to optimized primer sequences, EpiDesigner delivers an easy-to-read graphical interpretation of the amplicons designed over your target regions, as well as annotating distinct CpG sites covered by the assays.
STEP 2: Bisulfite Treatment
Genomic DNA (gDNA) is treated with bisulfite which leads to a conversion of all unmethylated cytosines, while methylated cytosines remain unaffected.
STEP 3: PCR, In vitro transcription, and RNA Cleavage Design
The EpiTYPER Assay starts with PCR using T7-promoter-tagged reverse primers to amplify the target regions while preserving the bisulfite-induced sequence changes.
After shrimp alkaline phosphatase treatment, in vitro transcription is performed, to discard unincorporated DNA nucleotides, the T7 promoter added during PCR is used to transcribe the PCR product from the reverse strand, yielding a single-stranded RNA product. This RNA product is specifically cleaved with RNase A at uracil residues.
STEP 4: Data Acquisition and analysis
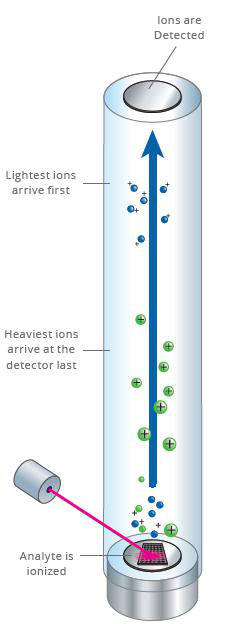
The EpiTYPER reaction products are dispensed onto a SpectroCHIP® Array (Chip). The Chip is then placed in the MALDI-TOF(Matrix Assisted Laser Desorption-Ionization Time of Flight) mass spectrometer for data acquisition, which typically requires 15-60 minutes. The results are automatically loaded into a database for data analysis with EpiTYPER software.
The matrix on the SpectroCHIP absorbs the energy of the laser and transfers it to the RNA fragments which subsequently become ionized. The ionized fragments are separated by the time it takes to arrive at the detector at the end of the mass spectrometer’s flight tube under the influence of an electric field. The time of flight increases with higher mass. A fragment containing one or more CpG dinucleotides is called a CpG unit. If a CpG dinucleotide was methylated and protected from bisulfite conversion, the corresponding RNA fragment, the CpG unit, will be 16Da heavier in mass when the CpG dinucleotide was methylated, resulting in a 16Da shift in the mass spectrum. The signal detected by the mass spectrometer for either fragment is proportional to the number of fragments. The number of fragments is quantified by the surface area of the corresponding peaks in the mass spectrum. The DNA methylation percentage of a given CpG is calculated by dividing the surface area of the peak representing the methylated fragment by the total surface area of the peaks of both the methylated and unmethylated fragment.
Key Features and Advantages
| Efficiency |
Precise & Accurate |
Sensitive |
Cost-Effective |
Simple Workflow |
- Bisulfite treated DNA to data in 8 hours
- Covers multiple CpGs in amplicons of up to 600 bp
- Compatible with formalin-fixed paraffin embedded tissue samples
|
- High precision (5% CV)
- High inter-laboratory reproducibility
|
- Detects down to 5% change in methylation levels
|
- 24-, 96-, and 384-well formats available
- Multiple CpGs analyzed in one simple reaction, from amplicon as long as 200-600 bp
|
- No need to design CpG-specific primers
- No PCR product purification required
- Ideal for investigating a few or several hundred target regions
- Convenient software solutions for comparison between samples
|
CD Genomics is now providing the efficient, cost-effective, accurate, and sensitive DNA bisulfate sequencing service by using EpiTYPER MassARRAY technology for your targeted DNA methylation studies. Please contact us for more information and a detailed quote.
For Research Use Only. Not for use in diagnostic procedures.


 Sample Submission Guidelines
Sample Submission Guidelines

