The Faster and Cheaper High-Throughput RNA Sequencing
April 19, 2019
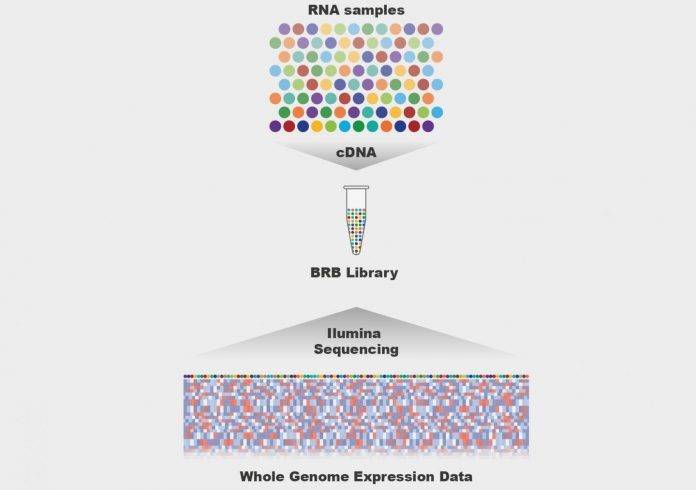 illustration of the BRB-seq method. [B. Deplancke/EPFL]
illustration of the BRB-seq method. [B. Deplancke/EPFL]
Genome-wide gene expression analyses by RNA sequencing (RNA-seq) have quickly become a standard in molecular biology because of the widespread availability of high throughput sequencing technologies. Nonetheless, RNA sequencing is still expensive and time-consuming, because it first requires the costly preparation of an entire genomic library—the DNA pool generated from the RNA of cells—while the data itself are also difficult to analyze. All this makes RNA sequencing difficult to run, rendering its adoption not as widespread as it could be.
Researchers from the lab of Bart Deplancke, PhD, at the École Polytechnique Federale de Lausanne (EPFL) Institute of Bioengineering say they have developed a new method called Bulk RNA Barcoding and sequencing (BRB-seq), which uses what is known as "sample barcoding" or "multiplexing". They show that BRB-seq is about 25 times less expensive than conventional commercial RNA sequencing technology, enabling the generation of ready to sequence libraries for up to 192 samples in a day with only 2 hours of hands-on time.
Among its many advantages, BRB-seq is quick and preserves strand-specificity, the scientists noted. As such, BRB-seq offers a low-cost approach for performing transcriptomics on hundreds of RNA samples, which can increase the number of biological replicates (and therefore experimental accuracy) in a single run, explained Deplancke, whose team’s work (“BRB-seq: ultra-affordable high-throughput transcriptomics enabled by bulk RNA barcoding and sequencing”) appears in Genome Biology.
In terms of performance, the scientists found that BRB-seq can detect the same number of genes as commercial technology at the same sequencing depth and that the technique produces reliable data even with low-quality RNA samples (down to RIN value = 2). Moreover, it generates genome-wide transcriptomic data at a cost that is comparable to profiling four genes using RT-qPCR, which is currently a standard, but low-throughput method for measuring gene expression, according to Deplancke. Therefore, they anticipate that BRB-seq or a comparable approach will over the longer term become standard in any molecular biology lab and replace RT-qPCR as the first gene expression profiling option.”
But researchers also said that there is still trouble with adapting and validating protocols for reliable and cheap profiling of bulk RNA samples—which is what they're faced with when trying to analyze the transcriptome of cells or tissues.


 Sample Submission Guidelines
Sample Submission Guidelines
