The Application of RNA Immunoprecipitation Sequencing (RIP-seq) in Hepatocellular Carcinoma (HCC) And Ovarian Cancer Research
RIP-seq (RNA immunoprecipitation) is a technology to study the binding of RNA and protein in cells. RIP uses the antibody of the target protein to precipitate the corresponding RNA-protein complex (RBP), isolate and purify the captured RNA, and combine the high-throughput sequencing technology to sequence and analyze the target RNA. RIP can be regarded as a similar application of chromatin immunoprecipitation ChIP technology, which binding target with protein is RNA and this technology has been widely used. RIP seq refers to high-throughput sequencing of enriched RNA fragments and is a powerful tool to understand the dynamic process of post transcriptional regulatory network, and also helps to discover RNA regulatory targets.
In recent years, RIP-seq has repeatedly made extraordinary contributions in the research of molecular mechanisms related to cancer pathology.
Experimental principle:
(1) Use antibodies or epitope markers to capture endogenous RNA-binding proteins in the nucleus or cytoplasm
(2) Prevent the binding of non-specific RNA
(3) The RNA binding protein and its binding RNA were separated by immunoprecipitation
(4) The combined RNA sequence was identified by microarray (RIP-chip), quantitative RT-PCR or high-throughput sequencing (RIP-Seq)
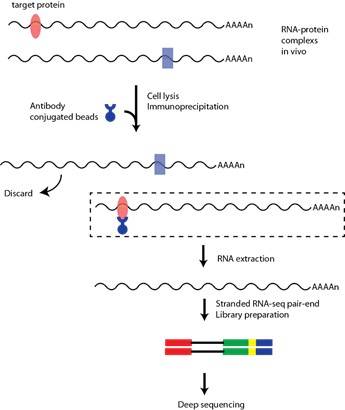 The experimental process of RIP-seq
The experimental process of RIP-seq
Application direction:
(1) Study the binding of RNA and protein in cells.
(2) Discover the interaction between RBP and non-coding RNA (LncRNA, miRNA, etc.).
(3) Mapping the genome-wide RNA and RBP interaction map.
RIP-seq helps to reveal the mechanism of protein post-translational modification
Hepatitis B virus (HBV) infection is a major risk factor for hepatocellular carcinoma (HCC). The RNA N6-methyladenosine (m6A) reader, YTH (YT521-B homology) domain 2 (YTHDF2), plays a critical role in the HCC progression. Recent studies found that YTHDF2 may be modified by O-GlcNAc and confirmed that Ser263 is the key site for O-GlcNAc modification of YTHDF2 through immunoprecipitation combined mass spectrometry (IP-MS). Additionally, the researchers screened and identified MCM2 and MCM5 as m6A modified molecules regulated by m6A-seq, RIP-seq, RNA-seq and other technologies. OGT-mediated YTHDF2 O-GlcNAcylation at Ser263 promoted the mRNA stabiliza-tion of MCM2 and MCM5 in an m6A-dependent manner in HCC. This study provides new ideas for finding new prognostic markers and molecular targets of HBV - associated hepatocellular carcinoma.
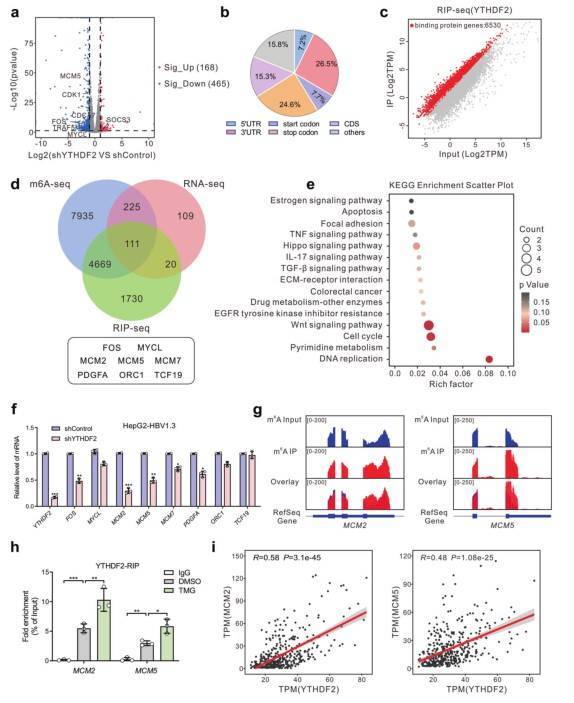 Identification of YTHDF2 targets by high-throughput RNA-seq, m6A-seq and RIP-seq
Identification of YTHDF2 targets by high-throughput RNA-seq, m6A-seq and RIP-seq
Identification of splicing events by RNA-seq and RIP-seq in ovarian cancer research
Alternative splicing has been found to be associated with various diseases including human cancers. Aberrant expression of splicing factors was found to promote tumorigenesis and the development of human malignant tumors. Ovarian cancer is the most lethal gynecologic malignancy and the abnormal expression of splicing factor is related to the development of ovarian cancer. Recent studies have found that the up-regulated proteins in HGSOCs are significantly enriched in splicing pathway proteins. The splicing factor USP39 is often over-expressed in HGSOC, and the increase of USP39 level is associated with poor prognosis. USP39 promotes proliferation/invasion in vitro and tumor growth in vivo and it was transcriptionally activated by the oncogene protein c-MYC in ovarian cancer cells concurrently. The variable splicing events regulated by USP39 were further identified by RNA sequencing (RNA-seq) and RNA immunoprecipitation sequencing (RIP-seq). Particularly, USP39 promotes the effective splicing of HMGA2 (high-mobility group AT-hook 2), thus increases the malignancy ofovarian cancer cells. This study indicates that USP39 may represent a new prognostic biomarker and a potential target of HGSOC.
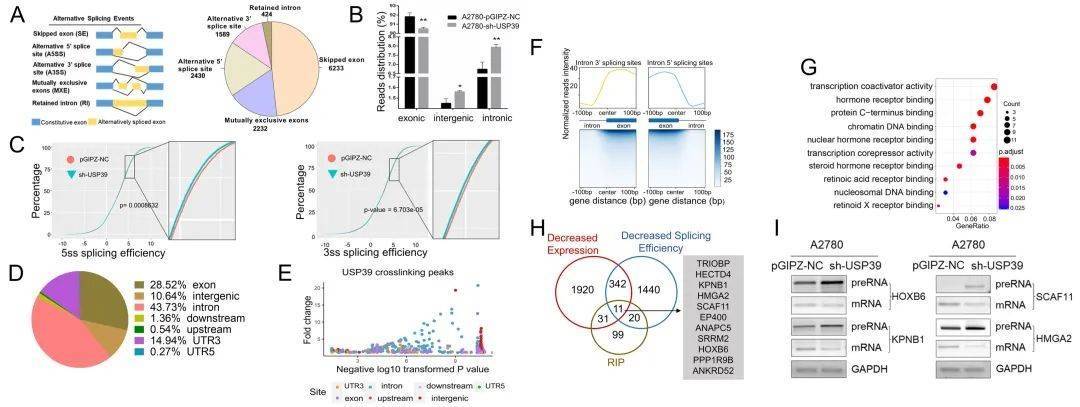 Identification of USP39-regulated splicing events by RNA-seq and RIP-seq
Identification of USP39-regulated splicing events by RNA-seq and RIP-seq
RIP technology is a powerful tool to study the dynamic process of post transcriptional regulatory network. The results detected by RIP-seq have wide coverage and high throughput. Its application can make the research level comprehensive and the mechanism more thorough and help us to understand RNA changes at the overall level of cancer and other diseases via high throughput.
References:
- Xie, Fei et al. "CircPTPRA blocks the recognition of RNA N6-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression." Molecular cancer vol. 20,1 68. 14 Apr. 2021.
- Wang, Shourong et al. "Splicing factor USP39 promotes ovarian cancer malignancy through maintaining efficient splicing of oncogenic HMGA2." Cell death & disease vol. 12,4 294. 17 Mar. 2021


 Sample Submission Guidelines
Sample Submission Guidelines
