Single-Cell Sequencing in Stress Research
Stress Research
Stress is a prevalent ecological factor that exerts a profound influence on various facets of organismal growth, development, metabolism, immunity, and adaptation. Consequently, the investigation of stress in animals and plants holds immense biological significance. For instance, in the realm of plant biology, stress profoundly affects multiple aspects of growth, development, and yield. When plants confront environmental stressors like drought, high temperature, low temperature, or salinity, they employ sophisticated regulatory mechanisms to modulate their physiological and metabolic states, enabling them to acclimate to changing surroundings. Moreover, stress can act as a catalyst for evolutionary processes within plant populations, driving the emergence of traits that enhance adaptability and survival. Consequently, the scientific scrutiny of stress provides invaluable insights into the adaptive mechanisms and evolutionary dynamics of organisms, ultimately serving as a foundational basis for the preservation and sustainable exploitation of biological resources.
Stress research has gained increasing significance in understanding the intricate relationship between psychological and physiological health. However, traditional research methods have often fallen short in capturing the complex cellular dynamics underlying stress responses. The emergence of single-cell sequencing has revolutionized the field, offering a powerful tool to delve into the unexplored territory of stress research.
Methods and Implications of Single-Cell Sequencing in Stress Research
Dissecting cell type and number changes
Single-cell sequencing allows researchers to identify changes in cell types and their abundance under stressful conditions. By analyzing the transcriptomic profiles of individual cells, researchers can determine which cell types are most affected by stress and understand the dynamics of cellular composition.
Uncovering the internal cellular response mechanisms
Single-cell sequencing reveals changes in signaling pathways, metabolic pathways, and gene expression profiles within individual cells. This enables researchers to unravel the intricate mechanisms by which cells respond and adapt to stressful environments, providing insights into cellular stress responses at a molecular level.
Discovery of novel stress response genes
Through single-cell sequencing, researchers can identify genes that are significantly upregulated or downregulated in response to stress. These genes may represent novel stress response genes that play essential roles in stress adaptation. Discovering and characterizing these genes contribute to a deeper understanding of stress response mechanisms.
Probing cell-to-cell interactions
Stress conditions can alter cellular interactions within tissues. Single-cell sequencing allows researchers to study the communication and interactions between different cell types under stressful environments. By elucidating the interplay between cells, researchers can gain insights into the mechanisms underlying cellular interactions during stress responses.
You might be interested in Cellular Differentiation and Development Research Based on Single-cell Sequencing.
A General Guide of Studying the Molecular Mechanism of Stress Using Single-Cell Sequencing
By delving into the cellular heterogeneity, gene expression patterns, and cellular interactions, single-cell sequencing enables a comprehensive exploration of the molecular landscape underlying stress responses.
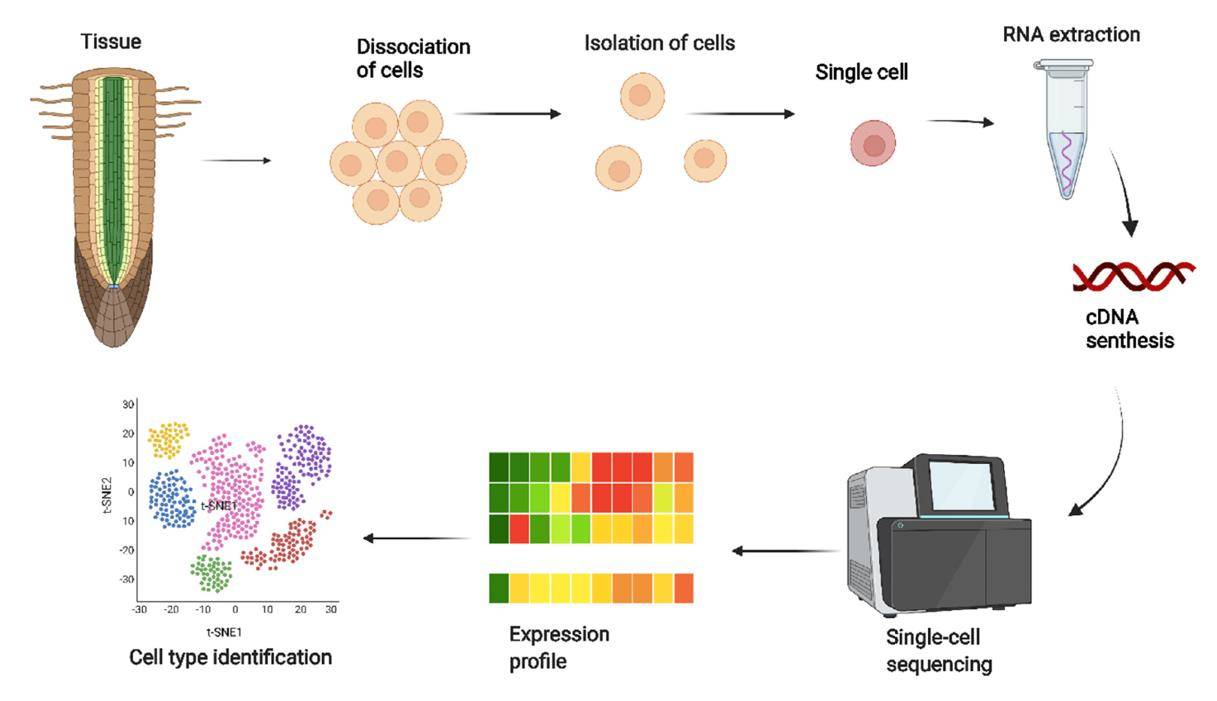 Single-cell RNA sequencing in Plant Research. (Bawa et al., 2022)
Single-cell RNA sequencing in Plant Research. (Bawa et al., 2022)
Experimental Design
- Define the specific type of stress you want to study and the phenotypic changes associated with it.
- Choose an appropriate stress model or system that mimics the stress condition you're interested in.
- Design experimental groups, including a control group and one or more stress groups based on the desired stress intensity or duration.
- Determine the time points for sample collection based on the dynamics of the stress response.
Phenotypic Characterization
- Assess the phenotypic changes induced by stress in the control and stress groups. This can involve behavioral observations, physiological measurements, or any other relevant phenotypic readouts.
- Determine the appropriate time points to capture the stress-induced phenotypic changes.
Sample Collection and Preparation
- Collect cells from your stress model at the desired time points.
- Perform dissociation of the tissue or organ to obtain single-cell suspensions.
- Preserve the cells carefully to maintain their RNA integrity during the dissociation process.
Single-Cell RNA Sequencing (scRNA-seq)
- Perform scRNA-seq to profile the gene expression of individual cells. There are several methods available, such as droplet-based techniques (e.g., 10x Genomics) or plate-based methods (e.g., Smart-seq2).
- Generate sequencing libraries from the isolated RNA of each cell, following the specific protocol for your chosen scRNA-seq method.
- Sequence the libraries using next-generation sequencing technologies.
Data Analysis
- Preprocess the raw sequencing data by removing adapters, filtering low-quality reads, and performing quality control.
- Align the reads to the reference genome or transcriptome to obtain gene expression counts for each cell.
- Normalize the gene expression data to account for differences in sequencing depth and other technical biases.
- Perform dimensionality reduction techniques (e.g., principal component analysis, t-SNE) to visualize the data and identify cell clusters.
- Identify differentially expressed genes (DEGs) between stressed and unstressed cells/clusters to pinpoint genes and pathways associated with the stress response.
- Conduct functional enrichment analysis to determine the biological processes, pathways, and gene ontology terms enriched in the DEGs.
- Explore the cellular heterogeneity and dynamics by analyzing the gene expression patterns and trajectories using specialized software or algorithms (e.g., Monocle, Seurat, Scanpy).
Validation and Follow-up Experiments
- Verify the findings from scRNA-seq by performing independent experiments such as qPCR, immunostaining, or Western blotting on specific genes or pathways of interest.
- Use additional techniques like in situ hybridization or immunohistochemistry to visualize the expression of specific genes or proteins in tissue sections.
Consider performing functional assays or perturbation experiments to investigate the functional consequences of specific genes or pathways identified in the scRNA-seq analysis.
Reference:
- Bawa, George, et al. "Single-cell RNA sequencing for plant research: Insights and possible benefits." International Journal of Molecular Sciences 23.9 (2022): 4497.


 Sample Submission Guidelines
Sample Submission Guidelines
