Exploring the Cell Cycle from A Single-Cell Transcriptome Perspective
Cell division is a fundamental biological process with features and basic mechanisms conserved in all eukaryotes. The process that occurs between the beginning of the growth of a new cell produced by cell division and the end of the next cell division to form daughter cells is usually referred to as the cell cycle. The cell cycle contains four phases: G1, S, G2, and M. It is a series of events in which cellular DNA replicates, grows, and divides.
Among tissues, even cells of the same type can be in different cell cycles. Traditional methods of cell cycle study, such as PI (propidium iodide) staining experiments and detection of related molecules, have disadvantages such as complicated experiments, limited available markers, more difficult to find intermediate state cells and difficult to analyze key genes in the process. In contrast, single-cell technology provides a new direction for cell cycle research because of its relative simplicity, the availability of a large number of gene expression profiles, the ability to capture rare cells, and the ability to find key genes without a priori information.
The Pseudotime Analysis in Cell Cycle Research
Cells begin to divide after a series of checkpoints for growth and division preparation. These steps are controlled by the activity of specific proteins that are regulated in time and space by transcriptional regulation, post-translational modifications, and protein degradation. Studies of the cell cycle have shown that hundreds of genes and proteins are regulated by the cell cycle. One of the advantages of single-cell RNA sequencing (scRNA-seq) is pseudotime analysis. Pseudotime analysis is used to infer the course of cellular changes from static data results by constructing trajectories of intercellular changes. In terms of specific classification analysis and complexity, pseudotime analysis can be divided into cell trajectory analysis and cell genealogy analysis.
Cell trajectory analysis refers to the change of cells along a specific direction, and the trajectory has a simpler starting and ending point; cell genealogy analysis usually refers to progenitor cells with multiple developmental trajectories and fates under specific conditions, and the change process resembles a complex tree-like structure. In addition, clustering analysis enables the reconstruction of continuous cell cycle processes. By analyzing specifically expressed genes, new genes with unknown associations with cell division and cell cycle can be identified, for example, other cell cycle-dependent (CCD) genes can be efficiently discovered using scRNA-seq, etc.
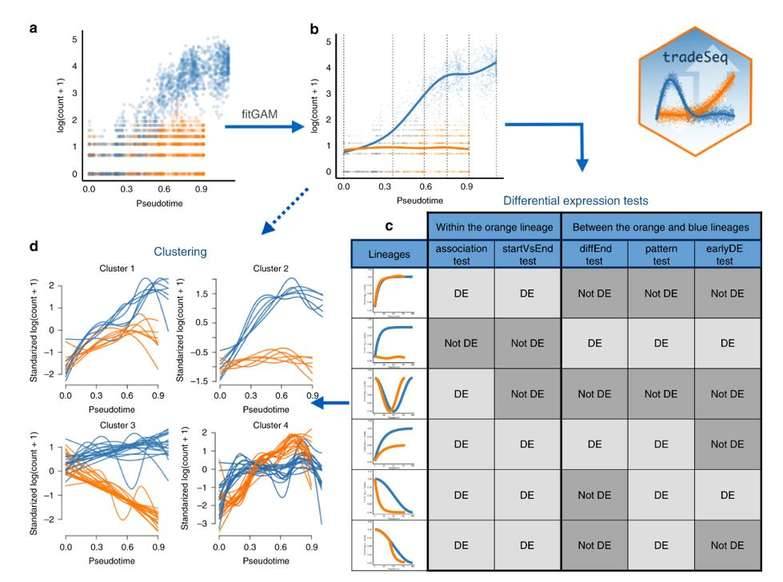 Trajectory-based differential expression analysis. (Van den Berge et al., 2020)
Trajectory-based differential expression analysis. (Van den Berge et al., 2020)
Except for scRNA-seq, cell cycle studies are usually performed in synchronized cell populations that are affected by gene expression, cell morphology and metabolic alterations, which limit the exploration of expression changes in cell cycle phases. Therefore, in addition to studies on developmental signaling regulation, molecular mechanisms, and resistance mechanisms, scRNA-seq has been widely used in cell cycle studies, e.g., classical questions such as biological trait development, questions on tissue regenerative capacity after trauma, and germ cell meiosis.
Comparative analysis of cell cycle-related genes can elucidate conserved and unique cell cycle-specific structural domains and also provide key information for functional analysis of cell cycle progression in single-cell RNA analysis and annotation of cell cycle marker genes. Single-cell transcriptome sequencing of important cell lines can also reveal cell cycle-dependent transcriptional heterogeneity present in cellular taxa. A continuous cell cycle progression was reconstructed by mimetic timing analysis, revealing differentiation trajectories including cell cycle entry and exit; a set of putative cell cycle-specific expressed genes was also identified.
Comparative analysis with cell cycle-related genes from other studies was performed to elucidate conserved and distinct stage-specific structural domains and genes. Cell cycle analysis correlated with mimetic analysis can be applied to indicate cell stemness, identify the effect of drugs to block cell cycle, etc., and has potential applications in differentiation and developmental processes, disease damage repair, and tumor immunogenesis.
The Annotation of the Cell Cycle
Each phase of the cell cycle has significant marker genes, and by analyzing the expression of genes related to the cell cycle, the cycle a cell is in can be annotated. Based on the cycle signature genes and the single-cell expression matrix, the cycle state that an individual cell is in can be evaluated to determine whether the cell is in a proliferative state.
 Cell cycle progression. (Wang 2021)
Cell cycle progression. (Wang 2021)
In meiosis studies, because germ cell meiosis in tissues does not occur simultaneously, it is difficult for the ordinary transcriptome to accurately reflect the mechanisms of gene expression regulation during meiosis, and the single-cell approach can well address the problem of heterogeneity. In single-cell transcriptome analysis, the cell cycle that each cell is in can be inferred from the data of gene clusters at specific cell cycle stages. For example, meiotic cells express several histone subunits with microtubule-binding proteins, and the combined expression of cell cycle proteins and cell cycle protein-dependent kinases does not appear in the original cells.
This phenomenon can be used to distinguish between the first stage of prophase meiosis and mitotic cell phase. The identification of primitive cell developmental regulatory genes by calculating the correlation of each gene with the cell cycle that most closely resembles the proposed developmental timing is an analysis that can distinguish primitive cell differentiation genes from cell cycle regulatory genes, which would not be possible using data from whole tissues. It can be seen that by studying the expression patterns of specific genes in individual cells, cells at different stages of the cycle can be effectively identified and categorized.
Cells in the same phase cluster with each other while cells in different phases are clearly separated and form a cycle in the distribution of PCA, which allows the dynamic process of the cell cycle to be inferred from static data, solving the difficulty of cell heterogeneity and data homogenization and allowing precise answers to scientific questions at the cellular level.
Impact of the Cell Cycle on Data Analysis
When performing cell cycle studies, tissues may contain two vastly different cell populations, e.g., containing cells that are proliferating and cells that are quiescent or senescent, and methods using molecular assays can only report an intermediate state, which then does not truly reflect either cell type.
When focusing on the heterogeneity of cell types, the cell cycle may be considered as the major systematic bias. In single-cell transcriptome data, cells of the same type often come from different cell cycle stages, which may affect downstream clustering analysis, manifesting as the division of the same type of cells into different cell populations. In addition, when performing cell clustering, different types of cells may be in a proliferative state and be clustered into groups because of high expression of cell cycle-related genes, which is actually a mixture of multiple cells. Therefore, the need to remove the effect of cell cycle on clustering should be considered during the analysis based on the understanding of the whole experiment.
Cell cycle analysis allows us to understand the state that the cells are in, and when the clustering results are heavily influenced by the cell cycle, the influence of the cell cycle needs to be removed based on the scientific question under study, leaving the biologically significant fraction. If the fraction is very small, it is also possible to choose not to treat it. In addition, combining different types of single-cell analysis, such as live cell imaging with fluorescent biosensors with single cell transcription or genomics, to link the molecular behavior of a cell to its subsequent fate may be an important direction for cell cycle research.
Conclusion
In general, single-cell studies can address the issue of cell heterogeneity. By single-cell transcriptome sequencing combined with clustering analysis and mimetic timing analysis, we can reconstruct the continuous cell cycle process and thus infer the cellular value-added state; by analyzing specifically expressed genes, we can discover known and unknown genes associated with cell division and cell cycle; by analyzing the molecular information of cells, we can infer the differentiation and developmental fate of cells, etc. It can be said that single-cell cell cycle studies have important application potential in developmental signaling regulation, disease damage repair, tumor immunogenesis, etc.
References:
- Van den Berge, Koen, et al. "Trajectory-based differential expression analysis for single-cell sequencing data." Nature communications 11.1 (2020): 1201.
- Wang, Zhixiang. "Regulation of cell cycle progression by growth factor-induced cell signaling." Cells 10.12 (2021): 3327.


 Sample Submission Guidelines
Sample Submission Guidelines
