We use cookies to understand how you use our site and to improve the overall user experience. This includes personalizing content and advertising. Read our Privacy Policy
Navigation
- Home
- Sequencing
- Genomics Sequencing
- Complete Plasmid/Phage Sequencing
- Mitochondrial DNA (mtDNA) Sequencing
- Viral Genome Sequencing
- Whole Genome Sequencing
- Whole Exome Sequencing
- Targeted Region Sequencing
- Amplicon Sequencing Services
- Chloroplast DNA (cpDNA) Sequencing
- TCR-Seq
- Shallow Whole Genome Sequencing
- Human Mitochondrial DNA (mtDNA) Sequencing
- Long-Read Sequencing
- ctDNA Sequencing Services
- Gene Panel Sequencing Service
- Long Amplicon Analysis (LAA)
- Animal/Plant Exome Sequencing Service
- Transcriptomics
- Bacterial RNA Sequencing
- RNA-Seq
- Small RNA Sequencing
- LncRNA Sequencing
- CircRNA Sequencing
- Ribosome Profiling (Ribo-seq)
- Total RNA Sequencing
- Targeted RNA Sequencing
- Degradome Sequencing
- Exosomal RNA Sequencing
- Ultra Low RNA Sequencing
- Dual RNA-seq
- microRNA Sequencing Service
- mRNA Sequencing Service
- Epigenomics
- Targeted Bisulfite Sequencing
- EM-seq Service
- Reduced Representation Bisulfite Sequencing
- Whole Genome Bisulfite Sequencing (WGBS)
- MeDIP Sequencing (MeDIP-Seq)
- ChIP-Seq
- MeRIP Sequencing (m6A Analysis)
- RIP-Seq
- ATAC-Seq
- NGS-BSP
- DNA 6mA Sequencing
- DAP-Seq Service (DNA affinity purification sequencing)
- 5mC/5hmC Sequencing
- oxBS-seq
- hMeDIP-seq
- Nanopore RNA Methylation Sequencing Service
- RNA Methylation Sequencing Service
- 2'-O-RNA Methylation Sequencing Service
- PacBio SMRT Sequencing
- Microbiome
- Genome Editing Sequencing
- Nanopore Sequencing
- Other Services
- Antibody Screening Sequencing (Phage Display Library Screening)
- Immune Repertoire Sequencing
- Sanger Sequencing
- Pre-made Library Sequencing
- Lentiviral/Retroviral Integration Sites Analysis
- AAV Genome Sequencing
- AAV (Adeno-Associated Virus) Integration Site Analysis
- 10x Spatial Transcriptome Sequencing Service
- HLA Typing
- Genomics Sequencing
- Genotyping
- Population Genetics
- Bioinformatics
- Microarray
- Applications
- Company
- Get Your Instant Quote


 Sample Submission Guidelines
Sample Submission Guidelines
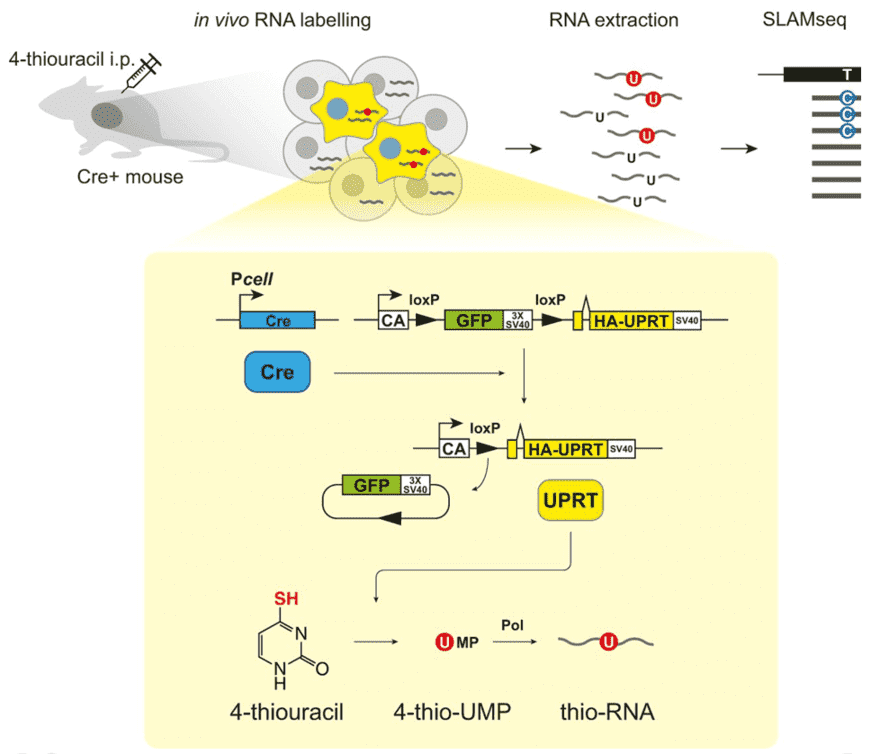 SLAM-ITseq design. Schematic of how SLAM-ITseq works. Cre is expressed in cells in which a cell type-specific promoter (Pcell) is activated and removes the floxed GFP-coding sequence followed by simian virus 40 polyadenylation sequence (SV40) between chicken β-actin promoter (CA) and UPRT-coding region, resulting in UPRT expression in the cells. When the mice are exposed to 4-thiouracil, only those UPRT-expressing cells (shown in yellow) can convert 4-thiouracil to 4-thio-UMP to synthesise thio-RNA. RNA is extracted from entire tissue without cell sorting, and the labelled RNA that is synthesised in the cells of interest is identified by finding T>C containing reads using SLAMseq.
[Figure/ Matsushima W , Herzog V A , Neumann T , et al. SLAM-ITseq: sequencing cell type-specific transcriptomes without cell sorting[J]. Development, 2018, 145(13):dev.164640.]
SLAM-ITseq design. Schematic of how SLAM-ITseq works. Cre is expressed in cells in which a cell type-specific promoter (Pcell) is activated and removes the floxed GFP-coding sequence followed by simian virus 40 polyadenylation sequence (SV40) between chicken β-actin promoter (CA) and UPRT-coding region, resulting in UPRT expression in the cells. When the mice are exposed to 4-thiouracil, only those UPRT-expressing cells (shown in yellow) can convert 4-thiouracil to 4-thio-UMP to synthesise thio-RNA. RNA is extracted from entire tissue without cell sorting, and the labelled RNA that is synthesised in the cells of interest is identified by finding T>C containing reads using SLAMseq.
[Figure/ Matsushima W , Herzog V A , Neumann T , et al. SLAM-ITseq: sequencing cell type-specific transcriptomes without cell sorting[J]. Development, 2018, 145(13):dev.164640.]