Sequencing Primers
What is sequencing primer?
Sequencing primers are short DNA oligonucleotides used in DNA sequencing methods. These primers provide a starting point for DNA synthesis during the sequencing reaction.
Sequencing primers in Sanger sequencing
In Sanger sequencing, which is also known as chain termination sequencing, sequencing primers are used to initiate DNA synthesis in a template DNA molecule. These primers are complementary to a specific region of the DNA template that needs to be sequenced. The sequencing reaction involves incorporating fluorescently labeled dideoxynucleotides (ddNTPs) into the growing DNA strand, which terminates the chain elongation at each position, resulting in a series of fragments of varying lengths. These fragments are then separated by size using capillary electrophoresis, and the sequence is determined based on the order of the terminated fragments.
Sequencing primers in NGS
In next-generation sequencing techniques, such as Illumina sequencing, sequencing primers are also used to initiate DNA synthesis. However, the sequencing process differs from Sanger sequencing. NGS platforms use massively parallel sequencing, where many sequencing reactions are carried out simultaneously in a high-throughput manner. Sequencing primers in NGS methods typically contain adapter sequences that are required for the attachment of the DNA fragments to the sequencing platform's solid surface or flow cell. These adapters facilitate the amplification and sequencing of the DNA fragments in a massively parallel manner, generating millions of short DNA sequences in a single sequencing run.
What are the principles of sequencing primer design?
Sequencing primers we often divided into vector universal sequencing primers and gene-specific sequencing primers:
Plasmid vector universal sequencing primers
Both ends of the MCS (polyclonal site), and then simply put, the sequencing primers inserted into the position of the ends of the fragment, that is, the primers designed according to the vector sequence, general sequencing companies have a part of free universal primers can be used, these primers are some common commercial vectors used sequencing primers, such as pCDNA3.1 vector, or pGL3- Basic vector, and of course other vectors.
General requirements for vector universal specificity primers:
(1) Specificity: requires only one binding site to the vector sequence;
(2) Position: at least 100 bases from the inserted fragment are required, too close may lead to inaccurate sequence sequencing at both ends of the inserted fragment
(3) Length: 17 to 25 bases are sufficient
(4) GC content: no special requirements, try to be above 35%
(5) Tm value: no special requirements, try to be above 40
(6) synthesis requirements: preferably PAGE purification, desalting is also possible, I myself use desalted primers without problems
Gene-specific sequencing primers
Generally, when the length of the inserted fragment exceeds 1500bp, we need to design gene-specific sequencing primers, how many need to design, that simply according to a sequencing primer effective read length is 700bp calculation, such as the inserted fragment of 2500bp, then at least four sequencing primers, two universal sequencing primers, two gene-specific sequencing primers.
General requirements for gene-specific sequencing primers:
(1) Specificity: binding to the sequencing template, with no mismatches of more than 4 bases
(2) Position: When more than one primer needs to be designed, the overlap between the sequencing length of the previous primer and the next sequencing primer is required to be at least 100 bases, otherwise the two sequencing primers may not lap, and the sequencing primer needs to be designed again. That is the same reason for vector universal primers and gene-specific primers
(3) Length: 17-25 bases
(4) GC content: no special requirement, try to be above 35%
(5) Tm value: no special requirement, try to be above 40
(6) Synthesis requirements: preferably PAGE purification, desalting is also possible
Sanger sequencing primer design
In Sanger sequencing experiments, most PCR primers can be used as sequencing primers for sequencing reactions, but some PCR primers cannot be used for sequencing reactions, mainly because of the following reasons:
(1) Simple primers: simple primers often have multiple binding sites on the sequencing template, which directly affects the sequencing results.
(2) Random primers: random primers (such as RAPD primers) are generally short, and the annealing temperature used is low, which cannot bind well to the template under the sequencing reaction conditions.
(3) Primers with too-long fragments: Generally, sequencing primers are required to be no larger than 24bp, and too-long primers are likely to bind to multiple binding sites present on the sequencing template under the lower sequencing reaction conditions, resulting in higher noise; in addition, the purity of longer primers will also be difficult to ensure, usually the purity of primers used for sequencing should be above 90%, and when the purity of primers is low, the background signal of the sequencing reaction will be significantly higher, which directly affects the accuracy of sequencing results.
(4) Specially labeled primers: all four bases in the Sanger sequencing reaction are fluorescently labeled, and fluorescently labeled primers used in the sequencing reaction will generate interference with the fluorescence signal; in addition, primers labeled with other types of macromolecules are also unsuitable for sequencing reactions, and the macromolecular groups labeled on the primers will directly affect the mobility of DNA fragments, resulting in poor or incorrect peak shape of sequencing results.
(5) Primers with low purity: sequencing primers have high requirements for purity, and mixed foreign substances in synthetic primers can lead to increased noise, which directly affects sequencing quality.
Primer selection in 16S/18S/ITS sequencing
Different species of bacteria have the same conserved region sequence and different variable region sequences. Primers can be designed to amplify all bacterial 16SrRNA genes in environmental samples based on the conserved region sequences, while different species of bacteria can be distinguished based on the variable region sequences.
The most commonly used primers in traditional methods are 27F and 1492R, which can amplify almost the full length of the complete 16SrRNA gene, which is not applicable to high-throughput sequencing platforms due to the current read length limitation of NGS, but is widely used for molecular identification of pure bacteria. Considering the current read length limitation of mainstream high-throughput sequencing platforms, only a certain variable region of 16SrDNA can be sequenced. Some choose to sequence single V region (V3/V4/V6), some double V region (V3-V4 region or V4-V5 region), and some choose triple V region (V1-V3 region, V5-V7 region or V7-V9 region) for 16S rDNA sequencing in the literature.
In general, the commonly used and more recognized sequencing regions for our environmental microbiomics are V3-V4, V4-V5, or V4 region alone. Many 16S sequencing primers are documented in the Earth Microbiome Project, including the traditional 515F/806R primers and optimized 515F/806R primer sequences.
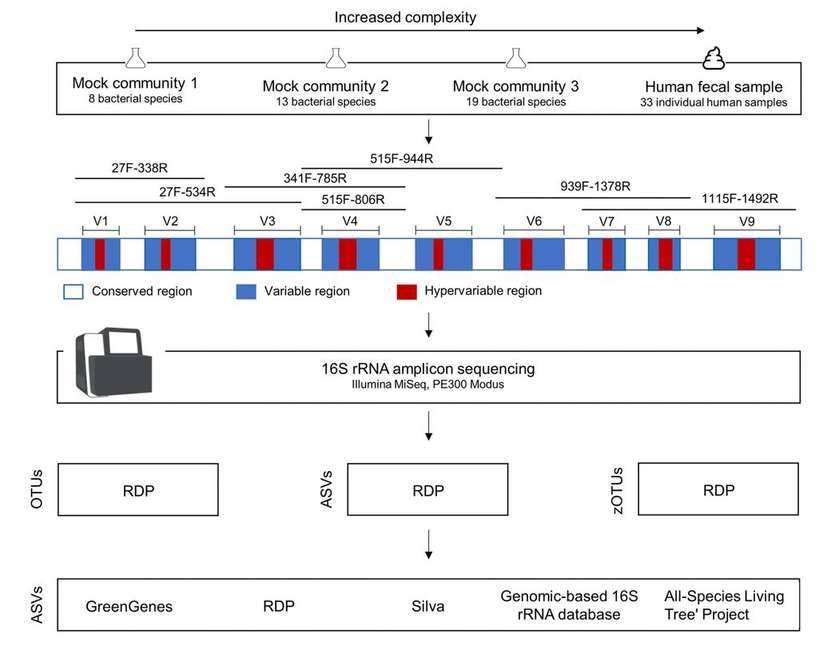 Primer in 16S rRNA gene sequencing. (Abellan-Schneyder et al., 2021)
Primer in 16S rRNA gene sequencing. (Abellan-Schneyder et al., 2021)
Similarly, 18S rRNA genes are DNA sequences that encode small subunits of eukaryotic ribosomes with both conserved and variable regions (V1-V9, no V6 region). The conserved regions reflect the affinities among biological species, while the variable regions reflect the differences among species and are suitable as taxonomic criteria at the species level and above. V4 is the most used choice for 18S rRNA gene analysis annotation, the most complete database information and the best classification. In fungi, 5.8S, 18S and 28S rRNA genes are highly conserved, while ITS shows extremely broad sequence polymorphism in the vast majority of eukaryotes due to less pressure from natural selection and the ability to tolerate more variation during evolution. At the same time, the conserved type of ITS shows relatively consistent within species and more obvious differences between species, which can reflect the differences between genera and even strains. The ITS sequence fragments are small (350 bp and 400 bp for ITS 1 and ITS 2, respectively) and easy to analyze, and have been widely used in the phylogenetic analysis of different species of fungi.
Archaebacteria, also known as archaea and archaea, are a very special class of bacteria that have some characteristics of both prokaryotes and eukaryotes. Studies have shown that primer pairs 519F/915R and 344F/915R have the best specificity for archaebacteria. There is a preference for primer pairs adapted to different platforms, and for Illumina MiSeq 2×300 bp sequencing platform, primer 519F/915R is the most suitable (16S V4V5 region).
Reference:
- Abellan-Schneyder, Isabel, et al. "Primer, pipelines, parameters: issues in 16S rRNA gene sequencing." Msphere 6.1 (2021): e01202-20.


 Sample Submission Guidelines
Sample Submission Guidelines
