Overview of Targeted Sequencing
What is Targeted Sequencing?
Targeted sequencing (tNGS) involves the precise enrichment of specific regions or loci within the genome, followed by sequencing using next-generation methods such as next-generation sequencing. This includes techniques like whole exome sequencing, which focuses on the protein-coding regions of the genome, as well as customized sequencing panels tailored for the investigation of particular genes of interest.
Recommended reading: WGS vs. WES vs. Targeted Sequencing Panels.
CD Genomics short-read sequencing and long-read sequencing platforms facilitate the robust analysis of exomes and genomes. This advanced targeted sequencing approach allows for comprehensive and efficient examination of genetic material, providing valuable insights into the molecular landscape and potential biomarkers associated with various conditions.
History of Targeted Sequencing
The inception of gene sequencing in life sciences research traces back to pivotal moments in scientific history:
In 1977, Walter Gilbert and Frederick Sanger pioneered the first sequencer, employing chain-termination sequencing to decode the genome sequence of phage X174, spanning 5,375 bases. This breakthrough marked the formal initiation of gene sequencing in scientific exploration.
Building on this foundation, in 1988, Chambehian et al. introduced multiplex PCR technology, laying the groundwork for subsequent advancements in multiplex PCR amplicon sequencing methodologies.
A significant milestone in the evolution of targeted sequencing occurred in 2005, when Nature Methods published an article titled "Direct Genomic Selection." This approach utilized 150kb biotin-labeled BAC DNA for hybridization with human genomic DNA, followed by the capture of DNA fragments using streptavidin affinity beads. Subsequent PCR amplification facilitated sequencing, revealing that approximately 50% of the sequenced fragments originated from the target region.
Today, targeted sequencing predominantly employs two methodologies: targeted capture and multiplex PCR, also known as amplicon sequencing.
Targeted Sequencing vs. Whole Genome Sequencing
Whole-genome sequencing (WGS) entails sequencing all bases of the entire genome, providing a comprehensive profile of the genome's sequence. Its primary applications include genome assembly and the identification of various genomic variants, including structural variants.
Targeted Sequencing (also known as gene panel sequencing) selectively sequences specific genes, typically ranging from a few dozen to a thousand genes. Therefore, in terms of genome coverage, whole genome sequencing > whole exome sequencing > targeted sequencing.
Whole-Exome Sequencing can be viewed as a subset of targeted sequencing, focusing solely on sequencing all exons within the genome.
WGS offers the most extensive detection scope, encompassing coding and non-coding regions, as well as regulatory regions and structural variants. But compared to targeted sequencing, WGS does have some limitations.
- Analysis and interpretation complexities arise from the detection of numerous introns and intragenic variants.
- Incurs higher detection costs compared to WES, particularly when ensuring data depth and quality.
- Requires complex methodologies for data storage, analysis, and annotation due to increased data volume.
- Lack of reference for rare variants in large samples may hinder pathogenicity analysis.
In comparison, targeted sequencing allows for deep sequencing due to its smaller detection region (e.g., exons comprise only 1% of all human gene sequences). This enables the detection of low-frequency and rare variants while reducing costs and storage requirements. Therefore, targeted sequencing offers a more cost-effective solution, especially for research endeavors with limited funding and studies focused on coding protein variant diseases.
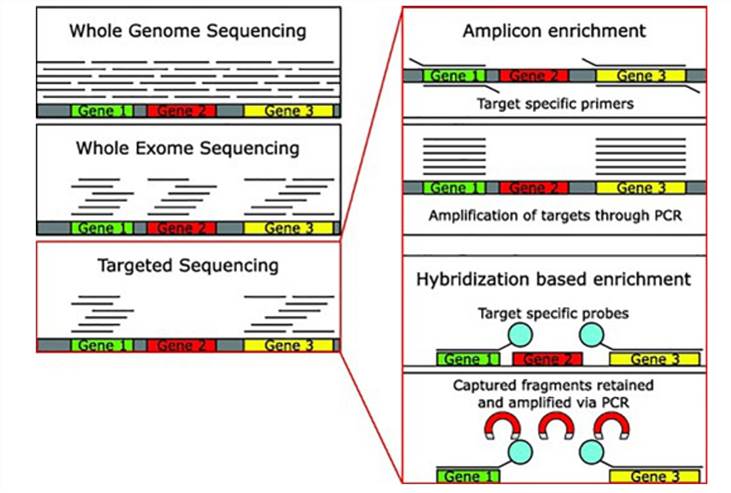 Methods of DNA-seq. (Bewicke-Copley et al., 2019)
Methods of DNA-seq. (Bewicke-Copley et al., 2019)
Hybridization Capture Targeted Sequencing
Hybridization capture stands as a targeted sequencing marvel, seamlessly melding molecular hybridization with next-generation sequencing techniques. This sophisticated method hinges on the meticulous design and synthesis of probes tailored to the target genomic region. These probes, acting as molecular magnets, selectively bind to the desired fragments within the target region, while extraneous segments are swiftly removed.
- Solid-Phase Hybridization: This variant entails the attachment of the probe to a solid-phase chip, facilitating the capture of the target region through precise probe-target interactions.
- Liquid-Phase Hybridization: In this iteration, the experimental process unfolds in a liquid milieu. Probes are equipped with biotin moieties, enabling their retrieval post-hybridization using streptavidin-coated magnetic beads. Upon completion of hybridization, these beads selectively trap the probe-bound target fragments. Subsequent elution rids the solution of unbound fragments, followed by denaturation to liberate the captured target fragments. Through a strategic magnetic capture, residual unbound probes are then removed, ensuring purity and completeness of the captured sequences.
Hybridization capture thus orchestrates a symphony of molecular interactions, culminating in the precise isolation and sequencing of target genomic regions.
Multiplex PCR Targeted Sequencing
Multiplex PCR targeted sequencing, also referred to as targeted amplicon sequencing, seamlessly integrates multiplex PCR technology with next-generation sequencing methods. This innovative approach enables simultaneous amplification of multiple target region sequences, yielding amplicon products. Subsequently, adapter sequences necessary for next-generation sequencing are introduced to both ends of the amplicon products, achieved through either PCR amplification or enzyme ligation reactions. This step transforms the amplicons into libraries ready for next-generation sequencing.
Following library preparation, next-generation sequencing is conducted, followed by an analysis of raw data. This comprehensive process yields sequence information specific to the target regions, fulfilling the primary objective of targeted sequencing.
- Common Applications
Multiplex PCR targeted sequencing finds widespread application in various fields. For instance, in the realm of pathogen detection, this technique facilitates the analysis of community composition and distribution of pathogenic microorganisms. Such analyses play a crucial role in diagnosing clinical pathogenic infections, contributing significantly to disease management and treatment strategies.
Multiplex PCR targeted sequencing thus emerges as a versatile tool, facilitating precise detection and characterization of target genomic regions across diverse applications.
Table 1 Differences between WGS, hybridization capture NGS panels, and multiplex PCR NGS panels
| Whole Genome Sequencing | Hybridization Capture (Exome Sequencing) | Multiplex PCR (Exome Sequencing) | |
| Target Region Size | 3 G (human) | 50 M | 10 kb - 5 M (variable) |
| Genome Coverage | 100% of the genome | 1.30% (variable) | <0.1% (variable) |
| Library Construction Cost | $ | $$ | $$ (variable) |
| Typical Sequencing Depth | 30x | 100x | 500-10,000x |
| Sequencing Data Volume | 90 Gb | 5 Gb | 1 Gb (variable) |
| Sequencing Cost | $$$ | $$ | $ (variable) |
| Data Storage Cost | $$$ | $$$ | $ |
| Difficulty in Analyzing Raw Data | High Complexity | Medium Complexity | Low Complexity |
Evaluating Data for Targeted Sequencing
The quality of data obtained from target gene region capture is primarily assessed through the following key indicators: target region coverage, capture efficiency, and homogeneity of target region coverage.
- Target Region Coverage: This metric denotes the proportion of detected regions compared to the total target region. Ideally, all regions of interest should be covered. However, due to probe design considerations such as GC content, sequence features, copy number variation, and sequence similarity, a small fraction of the target region may be excluded from capture, typically around 0-3%. Generally, higher target coverage indicates better probe or multiplex PCR performance.
- Capture Efficiency: This measure represents the percentage of data aligned to the target region out of the total data obtained. Higher capture efficiency reflects a greater utilization of sequencing data. It's crucial to assess sequence characteristics when designing probes. Probes that frequently bind to duplicate or high-copy sequence regions may capture non-target regions, thus reducing capture efficiency. Designing more specific probes can mitigate binding to non-specific sequences, thereby enhancing capture efficiency.
Evaluating these parameters ensures the reliability and effectiveness of targeted sequencing data, providing valuable insights into the intended genomic regions.
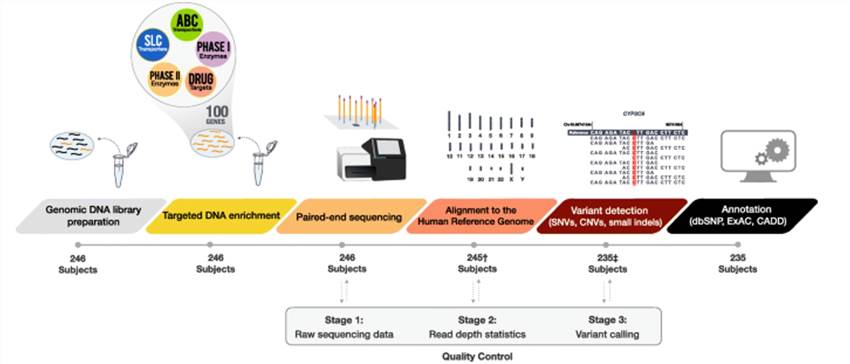 Targeted next generation sequencing sample and data processing workflow. (Gulilat et al., 2019)
Targeted next generation sequencing sample and data processing workflow. (Gulilat et al., 2019)
Factors Influencing Capture Efficiency
- High GC Regions: Regions such as untranslated regions (UTRs) and promoters often exhibit high GC content, leading to lower capture efficiencies. This discrepancy in capture efficiency between high GC regions and others can result in uneven coverage.
- DNA Quality: Input DNA quality significantly impacts capture bias, particularly evident in samples extracted from formalin-fixed paraffin-embedded (FFPE) tissues where fragmentation levels vary. Utilizing Agilent automated electrophoresis instruments like the 2100 Bioanalyzer or Tapestation analyzer for quality control ensures balanced capture and minimizes downstream analysis bias.
- DNA Input Quantity: Insufficient DNA input necessitates additional PCR cycles during library construction, increasing PCR duplicates and diminishing data utility. Advances in technology have reduced the required DNA input from micrograms to nanograms for targeted sequencing.
- Pseudogenes: Presence of pseudogenes can disrupt coverage uniformity, affecting data accuracy.
- DNA Fragment Size: Optimal matching of fragment size to probe design enhances capture efficiency. Utilizing Agilent automated electrophoresis instruments like the 2100 Bioanalyzer ensures accurate fragment size detection.
- Repeat Elements: Repeat elements can hinder reads distribution uniformity within the exome, necessitating increased sequencing depth for SNP detection.
- Coverage Homogeneity: Ensuring uniform depth of coverage across regions is vital for high-quality data. Achieving high homogeneity coverage involves careful pre-library construction. Methods like unbiased DNA fragmentation, employing amplification enzymes with low GC content preference, minimizing PCR enrichment cycles, and optimizing probe design and hybridization conditions enhance coverage homogeneity during capture experiments.
Applications of Targeted Sequencing
Targeted sequencing serves as a valuable complement to whole genome sequencing, streamlining both experimental procedures and analytical objectives. Its rapid and effective nature has carved out a unique niche in next-generation high-throughput sequencing, with a growing array of application areas.
- SNP Typing Assay: Targeted sequencing facilitates single nucleotide polymorphism (SNP) typing assays, enabling precise genetic variation analysis.
- Whole Exome Sequencing: By focusing on protein-coding regions, whole exome sequencing with targeted approaches offers insights into genetic variation associated with diseases and traits.
- Line Identification: Targeted sequencing aids in the identification and characterization of specific genetic lines, crucial in breeding and genetic studies.
- Population Heritability Analysis: Studying genetic variations across populations helps in understanding heritability and genetic diversity, vital for population genetics and evolutionary studies.
- Leaf Organoid/Mitochondrial Genome Sequencing: Targeted sequencing of specific organelle genomes or tissue-specific regions provides valuable information for understanding organoid development or mitochondrial genetics.
- Capture Technology: Targeted capture technologies enable precise sequencing of specific genomic regions, offering flexibility and efficiency in experimental design.
- Dominant Trait Screening: Targeted sequencing facilitates the identification of genetic variants underlying dominant traits, aiding in trait mapping and genetic improvement efforts.
- QTL Localization: Quantitative Trait Locus (QTL) mapping using targeted sequencing helps pinpoint genomic regions associated with complex traits, enhancing our understanding of genotype-phenotype relationships.
- Development of Molecular Markers for Breeding: Targeted sequencing contributes to the development of molecular markers for breeding programs, enabling marker-assisted selection and accelerated crop improvement.
- Methylation Sequencing: Targeted sequencing allows for precise analysis of DNA methylation patterns, offering insights into epigenetic regulation and gene expression dynamics.
- Species Classification, Phylogenetic Development: Targeted sequencing aids in species classification and phylogenetic studies by examining specific genomic regions or markers, shedding light on evolutionary relationships and biodiversity.
References:
- Bewicke-Copley, Findlay, et al. "Applications and analysis of targeted genomic sequencing in cancer studies." Computational and structural biotechnology journal 17 (2019): 1348-1359.
- Gulilat, Markus, et al. "Targeted next generation sequencing as a tool for precision medicine." BMC medical genomics 12 (2019): 1-17.


 Sample Submission Guidelines
Sample Submission Guidelines
