Exploring miRNA Sequencing Applications
As the field of miRNA sequencing progresses, numerous future avenues emerge, holding promise for advancing our comprehension of miRNA biology and its clinical ramifications. microRNAs (miRNAs) represent a class of small non-coding RNAs pivotal in post-transcriptional gene regulation, exerting substantial influence over diverse biological processes encompassing development, cellular differentiation, and the pathogenesis of diseases. The advent of miRNA sequencing stands as a transformative milestone, substantially enhancing our capacity to comprehensively delineate miRNA expression profiles, identify hitherto unrecognized miRNA entities, and unravel their functional importance in both physiological and pathological contexts. Within the confines of this discourse, we undertake a comprehensive exploration of the manifold applications of miRNA sequencing spanning various domains of scientific inquiry as well as clinical milieus.
Biomarker Discovery
A pivotal application of miRNA sequencing lies in the realm of biomarker discovery, wherein researchers endeavor to pinpoint miRNAs intricately linked with distinct diseases or physiological conditions. Through the meticulous profiling of miRNA expression within afflicted versus healthy tissues or biofluids, investigators can discern differentially expressed miRNAs, thereby unearthing prospective candidates for diagnostic, prognostic, or therapeutic intervention. Illustratively, discernible miRNA expression patterns have been delineated across a spectrum of maladies including cancers, cardiovascular afflictions, neurological ailments, and infectious diseases, furnishing invaluable insights into underlying pathogenic mechanisms and affording potential biomarkers for the early detection or surveillance of disease progression.
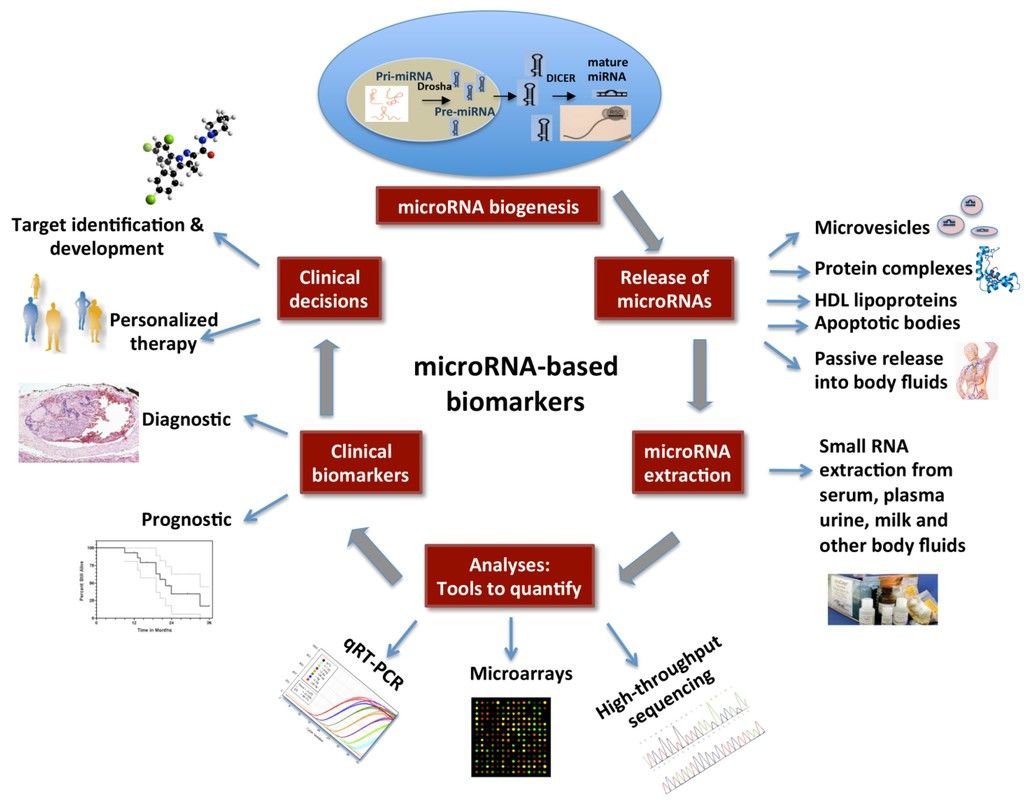 Circulating microRNAs as biomarkers
Circulating microRNAs as biomarkers
Functional Annotation of miRNAs
The advent of miRNA sequencing empowers researchers with a tool to unravel the functional intricacies of miRNAs, shedding light on their roles by pinpointing target genes and regulatory pathways. Through the integration of miRNA expression profiles with mRNA transcriptomic datasets, a nuanced understanding of miRNA-mRNA interaction networks emerges, elucidating downstream effects on gene expression. The functional annotation of miRNAs further enriches our comprehension, delineating their involvement in specific biological processes, signaling cascades, and disease phenotypes. Noteworthy is the discernible impact of miRNA-mediated regulation on pivotal signaling pathways such as Wnt, Notch, and TGF-beta, implicated across a spectrum of maladies spanning cancer, neurodegenerative disorders, and metabolic syndromes.
Characterization of miRNA Isoforms and Variants
The advent of miRNA sequencing heralds a profound opportunity for the comprehensive characterization of miRNA isoforms, known as isomiRs, and sequence variants, thereby providing intricate insights into miRNA biogenesis, maturation, and functional diversity. IsomiRs represent a spectrum of miRNA variants distinguished by differences in length, sequence, or post-transcriptional modifications, engendering variations in target specificity and regulatory functions. Through the meticulous profiling of miRNA isoforms, researchers unravel novel regulatory mechanisms and discern functional implications intertwined with miRNA diversity. Furthermore, the identification of single nucleotide polymorphisms (SNPs) and sequence variants within miRNAs or their target sites emerges as a critical avenue for elucidating their involvement in disease susceptibility, drug response modulation, and the realm of personalized medicine.
Cell-Type-Specific Expression Profiling
The technology of miRNA sequencing can be leveraged for profiling expression that is specific to various cell types, bestowing researchers with the potential to deconstruct the complexities, alteration, and dynamisms inherent to miRNA expression across diversely differentiated cell types, tissues, and stages of maturation. Employing modern single-cell miRNA sequencing technologies offer unparalleled resolution, thus enabling the dissection of discriminatory expression patterns of miRNA uniquely exhibited by individual cells. This permits the revelation of distinctive signatures and intricate networks of regulation specific to each cell type. Assessment of miRNA expression profiles that are cell-type-specific can provide critical insights into the mechanisms of cellular differentiation, the determination of various cellular lineages, and the communication that transpires between cells, all of which can be studied in the context of normal physiological conditions as well as in disease states.
Environmental and Epigenetic Influences on miRNA Expression
miRNA sequencing represents a potent tool in elucidating the intricate influence of environmental stimuli, epigenetic alterations, and genetic variances upon miRNA expression dynamics. Environmental stressors, encompassing pollutants, toxins, dietary constituents, and lifestyle determinants, wield the capacity to finely modulate miRNA expression profiles, thereby exerting substantial impacts on disease predisposition and pathogenic cascades. Concurrently, epigenetic modifications, typified by DNA methylation, histone post-translational modifications, and chromatin conformational changes, intricately orchestrate the regulation of miRNA expression levels and functionality.
The amalgamation of miRNA sequencing datasets with comprehensive epigenomic profiles and environmental repositories facilitates a holistic comprehension of gene-environment interplays, thus providing profound insights into their ramifications for human health and disease susceptibility.
Clinical Applications and Translational Research
miRNA sequencing harbours immense potential for clinical implementation and the facilitation of translational research, by providing possible pathways for disease diagnosis, prognosis, and therapeutic intervention. Identified through sequencing studies, miRNA biomarkers can be exploited for a range of non-invasive diagnostic procedures, such as the stratification of patients and the monitoring of treatment responses. Furthermore, therapeutic strategies that target aberrant miRNAs, including the use of miRNA mimics, antagomiRs, and small molecule inhibitors, are currently under investigation as a new frontier in precision medicine approaches. As it stands, clinical trials are determining the safety and efficacy of miRNA-based therapeutics for several diseases, encompassing cancer, cardiovascular disorders, and inflammation-based conditions.
Service you may intersted in
Learn more
Complete Overview of Small RNA Sequencing: Methods, Workflow, Platform, and Applications
Integration with Multi-Omics Data
Integration of miRNA sequencing with multifarious omics datasets, encompassing transcriptomics, genomics, epigenomics, and proteomics, empowers researchers to garner a more comprehensive understanding of gene regulatory networks. This provides profound insights into the intricate molecular mechanisms that dictate biological functions and disease pathogenesis.
The amalgamation of multi-omics data facilitates the discovery of convoluted interactions and regulatory inter-dependencies betwixt various molecular layers. This can illuminate an elaborate spectrum of disease mechanisms and present prospective therapeutic targets for rigorous investigation.
An exemplary application of this approach is the integrative analysis of miRNA sequencing and mRNA expression data. This combination can help identify miRNA-mRNA regulatory modules, neuralgic transcriptional regulatory networks, and aberrant pathways linked to disease-specific phenotypes.
Similarly, the merger of miRNA sequencing data with genomic and epigenomic data can expedite the identification of rare genetic variants, pivotal epigenetic alterations, and chromatin states that have consequential impacts on miRNA expression and functional activity.
Additionally, the integration of miRNA sequencing with proteomic data can shed light on the enigmatic realms of miRNA-mediated post-transcriptional regulation of protein expression. This can provide unprecedented insights into the consequential alterations in cellular phenotypes.
In conclusion, the incorporation of multi-omics data into miRNA research can significantly enhance the depth and breadth of our knowledge landscape. By offering a holistic perspective on molecular interactions and pathways, such integration can better elucidate the drivers underpinning both biological processes and disease states.
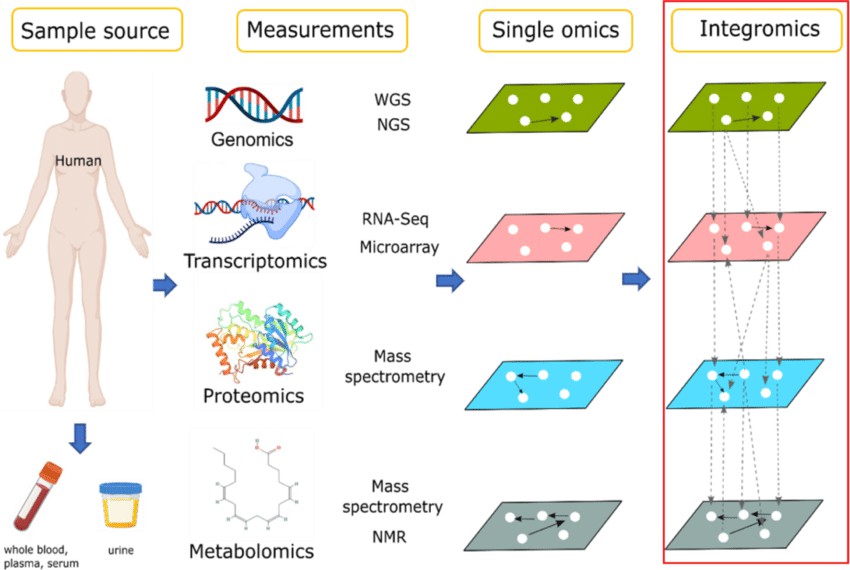 Multi-omics approach to the discovery of monitoring biomarker
Multi-omics approach to the discovery of monitoring biomarker
Longitudinal and Time-Series Analysis
miRNA sequencing holds considerable promise for conducting longitudinal and time-series analyses aimed at exploring the temporal dynamics of miRNA expression and regulatory networks. Longitudinal investigations entail the repeated assessment of miRNA expression within the same individuals or biological samples at multiple time points. This approach permits the monitoring of fluctuations in miRNA profiles over time and the identification of responsive changes to environmental stimuli, disease progression, or therapeutic interventions.
Conversely, time-series analysis involves the systematic profiling of miRNA expression across sequential time points within experimental systems or model organisms. By doing so, researchers can capture the evolving patterns of miRNA expression and regulatory events throughout biological processes such as development, differentiation, and response to stimuli.
The utilization of longitudinal and time-series analysis methodologies facilitates the elucidation of pivotal miRNAs that underlie dynamic alterations in gene expression, regulatory networks, and cellular phenotypes over time. Consequently, such approaches offer valuable insights into the temporal orchestration of biological processes and the trajectories of diseases.
Single-Cell and Spatially Resolved Analysis
Advancements in single-cell and spatially resolved sequencing technologies represent pivotal milestones in miRNA research, greatly expanding our capacity to explore the intricacies of miRNA expression within heterogeneous tissues and spatially defined compartments. Single-cell miRNA sequencing, for instance, empowers researchers to unravel the heterogeneity inherent in complex tissues and cell populations. This technique facilitates the identification of rare cell types, delineation of cell-state transitions, and characterization of cell-specific miRNA expression profiles.
Similarly, spatially resolved miRNA sequencing methodologies, such as spatial transcriptomics and spatially resolved RNA sequencing, provide avenues for mapping miRNA expression patterns within intact tissue sections. By doing so, these techniques unveil the spatial organization of miRNA expression, shed light on cellular interactions, and elucidate the microenvironmental influences shaping miRNA function.
The integration of single-cell and spatially resolved miRNA sequencing offers unparalleled insights into various aspects of cellular biology, including cell-to-cell communication, spatially constrained regulatory networks, and alterations in tissue architecture associated with disease. Consequently, these methodologies open new frontiers for understanding tissue physiology and unraveling the complexities of disease pathology at unprecedented resolution.
Machine Learning and Predictive Modeling
The application of machine learning and predictive modeling methodologies to miRNA sequencing data unveils intricate patterns, promotes the prediction of regulatory interactions, and categorizes samples on the basis of miRNA expression profiles. Supervised learning algorithms, inclusive of support vector machines, random forests, and neural networks, can be trained on miRNA sequencing datasets to classify samples into distinct disease subcategories, project clinical outcomes, and pinpoint molecular signatures corresponding to specific phenotypes. Machine learning models can also predict potential target genes, rate possible miRNA-target interactions, and disclose miRNA-mediated regulatory networks. The amalgamation of machine learning with miRNA sequencing analysis amplifies the interpretability, predictive capacity, and reproducibility of outcomes, thus fostering a data-driven discovery and the generation of hypotheses in miRNA research.
Therapeutic Target Identification
In the realm of precision medicine, miRNA sequencing holds immense potential for the discovery of innovative therapeutic targets. By revealing aberrant miRNA expression profiles characteristic of various pathological conditions, researchers are empowered to put forward hypothetical miRNAs with plausible involvements in disease onset, progression, and responses to treatment. With the direct therapeutic intervention of these dysregulated miRNAs using miRNA mimetics or inhibitors, potential treatment avenues for diseases as diverse as cancer, cardiovascular anomalies, neurodegenerative disorders, and infectious diseases emerge.
Furthermore, the technological leaps made in delivery systems based on nanoparticles, as well as advancements in CRISPR/Cas9 genome editing mechanisms, allow a uniquely aimed delivery of miRNA therapeutics to specific tissues or cellular subtypes, thereby reducing the likelihood of off-target repercussions and augmenting therapeutic effectiveness. This development underscores the importance of miRNA recognition and manipulation in unfolding the future of molecular disease intervention.
Drug Discovery and Development
The sequencing of miRNA offers indispensible insight into the molecular mechanisms underlying drug response and resistance, thereby streamlining the processes of drug discovery and development. By profiling the expression patterns of miRNA in the wake of pharmacological treatments, researchers can pinpoint miRNAs tied to either drug sensitivity or resistance phenotypes, clarify drug-target interactions, and prioritize the list of candidate miRNAs up for therapeutic modulation.
The incorporation of miRNA sequencing with drug screening assays, high-throughput compound libraries and sophisticated computational modeling methodologies can expedite the discovery of innovative drug candidates and facilitate the repositioning of extant drugs for additional therapeutic applications. Moreover, miRNA biomarkers distinguished via sequencing studies can serve as useful pharmacodynamic markers during clinical trials, enabling healthcare professionals involved to monitor drug efficacy, forecast the result of treatments and categorize patient groups for tailor-made therapy.
Regenerative Medicine and Stem Cell Therapy
miRNA sequencing is also considered instrumental in regenerative medicine and stem-cell therapy by revealing the regulatory mechanisms that dictate stem cell fate, differentiation, and tissue regeneration. The profiling of miRNA expression patterns throughout cellular reprogramming, differentiation, and organogenesis, enables a deeper understanding of the molecular pathways and epigenetic regulators that control stem cell pluripotency and lineage determination.
Furthermore, miRNA sequencing facilitates the recognition of lineage-specific miRNA signatures related to differentiating cell types, thereby bolstering the generation of functional tissues and organs for transplants and regenerative therapies. The specific manipulation of miRNAs expression, using miRNA mimics or inhibitors, optimizes the accuracy and efficacy of cellular reprogramming and tissue engineering methods, presenting newfound treatment pathways for degenerative conditions, tissue injuries, and inborn disorders.
Environmental and Toxicological Applications
As a crucial tool for environmental and toxicological studies, miRNA sequencing facilitates our understanding of the effects of pollutants, toxins, and environmental exposure on miRNA expression and its regulatory networks. By examining miRNA expression patterns under the influence of environmental stressors, chemicals, and drugs, we gain valuable insights into the molecular complexities underpinning toxicity, cellular adaptations, and stress responses.
Differential expression analysis of miRNAs in environmental samples - such as particulate content in air, contaminants in water, or additives in food - allows for the recognition of miRNA biomarkers that denote environmental pollution, human exposure levels, and consequent health risks. Furthermore, the implementation of miRNA sequencing serves a dual purpose, assessing both the efficacy and safety of environmental reclamation strategies, and examining the toxicity of industrial chemicals and pharmaceutical products. These insights deliver critical data that drive regulatory decisions, contributing to the safeguarding of environmental integrity and public health.
Conclusion
In culmination, microRNA sequencing emerges as a potent and multi-faceted instrument for dissecting the intricate matrix of miRNA expression and governance in both health and pathological conditions. The wide-ranging applications of miRNA sequencing encompass the discovery of biomarkers, functional elucidation, isoform profiling, cell-type-specific mapping, environmental impact analysis, and clinical implementation. By leveraging the potential of miRNA sequencing methodologies, researchers can unveil the convolutions of miRNA biology, thereby charting the route towards ground-breaking diagnostic tools, therapeutic interventions, and precision medicine approaches. As the field of miRNA research continues to advance, it engenders immense hope for enhancing our grip on gene regulation networks and ameliorating human health outcomes.
References:
- Khamina K, Diendorfer AB, Skalicky S, Weigl M, Pultar M, Krammer TL, Fournier CA, Schofield AL, Otto C, Smith AT, Buchtele N, Schoergenhofer C, Jilma B, Frank BJH, Hofstaetter JG, Grillari R, Grillari J, Ruprecht K, Goldring CE, Rehrauer H, Glaab WE, Hackl M. A MicroRNA Next-Generation-Sequencing Discovery Assay (miND) for Genome-Scale Analysis and Absolute Quantitation of Circulating MicroRNA Biomarkers. Int J Mol Sci. 2022
- Potla P, Ali SA, Kapoor M. A bioinformatics approach to microRNA-sequencing analysis. Osteoarthr Cartil Open. 2020 Dec 19;3(1):100131. doi: 10.1016/j.ocarto.2020
- Pérez-Rodríguez D, López-Fernández H, Agís-Balboa RC. Application of miRNA-seq in neuropsychiatry: A methodological perspective. Comput Biol Med. 2021 Aug;135:104603. doi: 10.1016/j.compbiomed.2021
- Oh JN, Son D, Choi KH, Hwang JY, Lee DK, Kim SH, Lee M, Jeong J, Choe GC, Lee CK. MicroRNA expression data of pluripotent and somatic cells and identification of cell type-specific MicroRNAs in pigs. Data Brief. 2020
- Motameny S, Wolters S, Nürnberg P, Schumacher B. Next Generation Sequencing of miRNAs - Strategies, Resources and Methods. Genes (Basel). 2010
- Sundarbose, K.; Kartha, R.V.; Subramanian, S. MicroRNAs as Biomarkers in Cancer. Diagnostics 2013, 3, 84-104.
- Pitaloka DAE, Syamsunarno MRAAA, Abdulah R, Chaidir L. Omics Biomarkers for Monitoring Tuberculosis Treatment: A Mini-Review of Recent Insights and Future Approaches. Infect Drug Resist. 2022


 Sample Submission Guidelines
Sample Submission Guidelines
