Navigating miRNA Sequencing: A Comprehensive Guide
Introduction
microRNAs (miRNAs), a class of small non-coding RNAs, are pivotal regulatory agents in gene expression, effectuating major changes post-transcriptionally. Their influence pervades diverse biological events, including cellular development, differentiation, and the pathogenesis of diseases. The ongoing unraveling of the complex labyrinth of regulatory networks orchestrated by miRNAs has necessitated the development of comprehensive and efficient strategies to punctiliously profile miRNA expression.
This surge of interest has led to the emergence of miRNA sequencing technology. Superseding traditional methodologies, this high-throughput approach serves as a powerful investigative apparatus, providing in-depth understanding into the intricate milieu of small RNA molecules. Robust and comprehensive, miRNA sequencing offers unprecedented insights into the miRNAome - a vibrant tapestry woven of countless miRNA sequences and their regulatory interactions. Exploring this enriches our understanding of the biological systems they regulate, paving the way for potential translational research in the field of molecular biology and medicine.
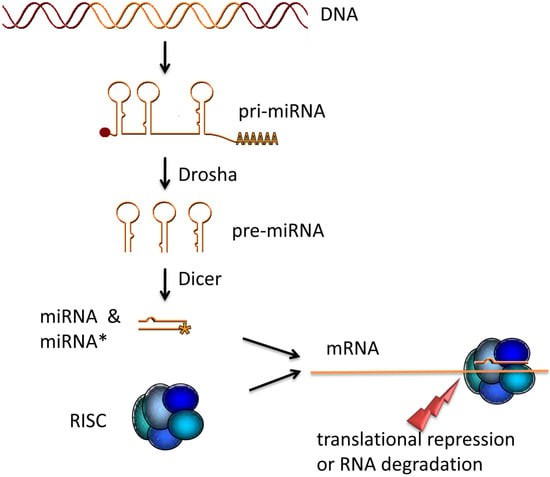 miRNA maturation
miRNA maturation
Overview of miRNA Sequencing
miRNAs are integral to post-transcriptional gene regulation, bearing significant influence over biological processes varying from development and differentiation to disease pathogenesis. As the depth of comprehension continues to expand concerning the complex regulatory networks governed by miRNAs, there is a growing requirement for comprehensive, high-throughput methodologies to precisely profile miRNA expression. Rising to this pressing demand, miRNA sequencing—also termed small RNA sequencing—has established itself as an instrumental tool. It is by far superior to its traditional counterparts, such as microarrays, as it extends beyond the constraints of pre-determined probes. miRNAs sequencing affords an unbiased and highly sensitive approach to the identification of known and hitherto undiscovered miRNAs. Capitalizing on high-throughput sequencing systems, it facilitates the researchers to decipher the full spectrum of miRNAs present in a sample. This in turn illuminates their varied functions and the nuances of their regulatory mechanisms, enriching our understanding of miRNAs landscape.
Service you may intersted in
Importance of Understanding miRNA Sequencing
The utility of miRNA sequencing extends across a wide array of biological and clinical research fields, reflecting a remarkable diversity in its applications. A prominent facet of its use lies in the sphere of biomarker discovery. Through the examination of variations in the expression of miRNAs in individuals during states of disease, as opposed to during health, it becomes possible to discern potential novel prognostic and diagnostic indicators.
Employing miRNA sequencing as a tool can also illuminate the regulatory influences of miRNAs in the pathogenesis of diseases. An integrative analysis that combines miRNA expression profiles with transcriptomic data regarding mRNA, allows researchers to decipher the complex interactions present between miRNAs and their corresponding target genes. Such insights yield invaluable information concerning the mechanisms of diseases and prospective therapeutic targets.
Moreover, the use of miRNA sequencing makes feasible the identification of previously undiscovered miRNAs, which could easily have been missed via traditional methodologies. This new population of miRNAs brims with potential, acting as potential biomarkers or therapeutic targets. Notably, they add to our comprehension of the labyrinthine nature of gene regulatory networks.
For more applications, refer to "miRNA Sequencing application".
Understanding the miRNA Sequencing Workflow
Overview of the Workflow Steps
The miRNA sequencing protocol conventionally encompasses four main stages: sample preparation, library construction, sequencing, and subsequent data analysis. In the initial phase of sample preparation, RNA molecules, inclusive of miRNAs, are isolated from the biological specimen and subsequently transformed into sequencing libraries. These libraries are subsequently subjected to high-throughput sequencing utilizing next-generation sequencing platforms, exemplified by Illumina systems. The resultant sequencing data undergo bioinformatic processing and analysis to quantify miRNA expression levels and discern differentially expressed miRNAs.
For specific workflow, please refer to "miRNA Sequencing Workflow "
Importance of Each Step in the Process
Every stage within the miRNA sequencing workflow is indispensable for achieving dependable and interpretable outcomes. Sample preparation is paramount for isolating RNA molecules of high quality, devoid of contaminants and degradation, thereby ensuring precise downstream analysis. Library construction entails adaptor ligation and amplification of miRNA sequences, facilitating their detection throughout sequencing. The sequencing phase yields millions of short reads delineating miRNA sequences, affording thorough coverage of the miRNA transcriptome. Lastly, data analysis encompasses bioinformatic processing of sequencing data to quantify miRNA expression levels, discern differentially expressed miRNAs, and unveil novel miRNAs.
Exploring miRNA Sequencing Analysis
miRNA sequencing analysis constitutes a multifaceted endeavor geared towards unraveling the intricate landscape of miRNA expression patterns and elucidating the regulatory networks underpinning biological processes and diseases. This analytical pursuit encompasses several pivotal components, each bearing a vital role in deciphering the nuanced functions of miRNAs in gene regulation.
Key Components of miRNA Sequencing Analysis
Quality Control and Pre-processing: The analysis commences with a meticulous evaluation of sequencing data quality to ensure the reliability of subsequent analyses. Critical quality control metrics, encompassing read quality scores, assessment for adapter contamination, and alignment rates, are scrutinized to identify and rectify potential sources of bias and variability. Pre-processing procedures, such as adapter trimming, quality filtration, and the removal of low-quality reads, are meticulously executed to augment the precision of ensuing analyses.
Alignment and Mapping: Subsequent to quality control procedures, sequencing reads undergo alignment and mapping against reference databases to ascertain the presence and expression levels of known miRNAs. Utilizing esteemed alignment algorithms like Bowtie and miRDeep2, reads are accurately mapped to miRNA sequences, facilitating the quantification of miRNA expression levels across diverse samples.
Molecular barcodes Handling: molecular barcodes are seamlessly integrated into the analytical pipeline to counteract amplification bias and refine the accuracy of miRNA quantification. Specialized molecular barcodes-tools are adeptly employed to process molecular barcodes-tagged reads, streamline duplicate identification, and derive molecular barcodes-corrected counts, thereby ensuring robust and reproducible quantification of miRNA expression levels.
Differential Expression Analysis: Rigorous comparative analyses are conducted to pinpoint differentially expressed miRNAs amidst varied experimental conditions or sample cohorts. Esteemed statistical methodologies such as DESeq2 and edgeR are harnessed to evaluate the significance of miRNA expression alterations and prioritize potential candidate miRNAs for further exploration.
Novel miRNA Discovery: Beyond the repertoire of known miRNAs, the miRNA sequencing analysis facilitates the discovery of novel miRNAs harboring potential regulatory roles. Leveraging sophisticated novel miRNA prediction algorithms alongside secondary structure prognostication and homology analysis, candidate novel miRNAs are discerned from the sequencing dataset with careful consideration for their regulatory implications.
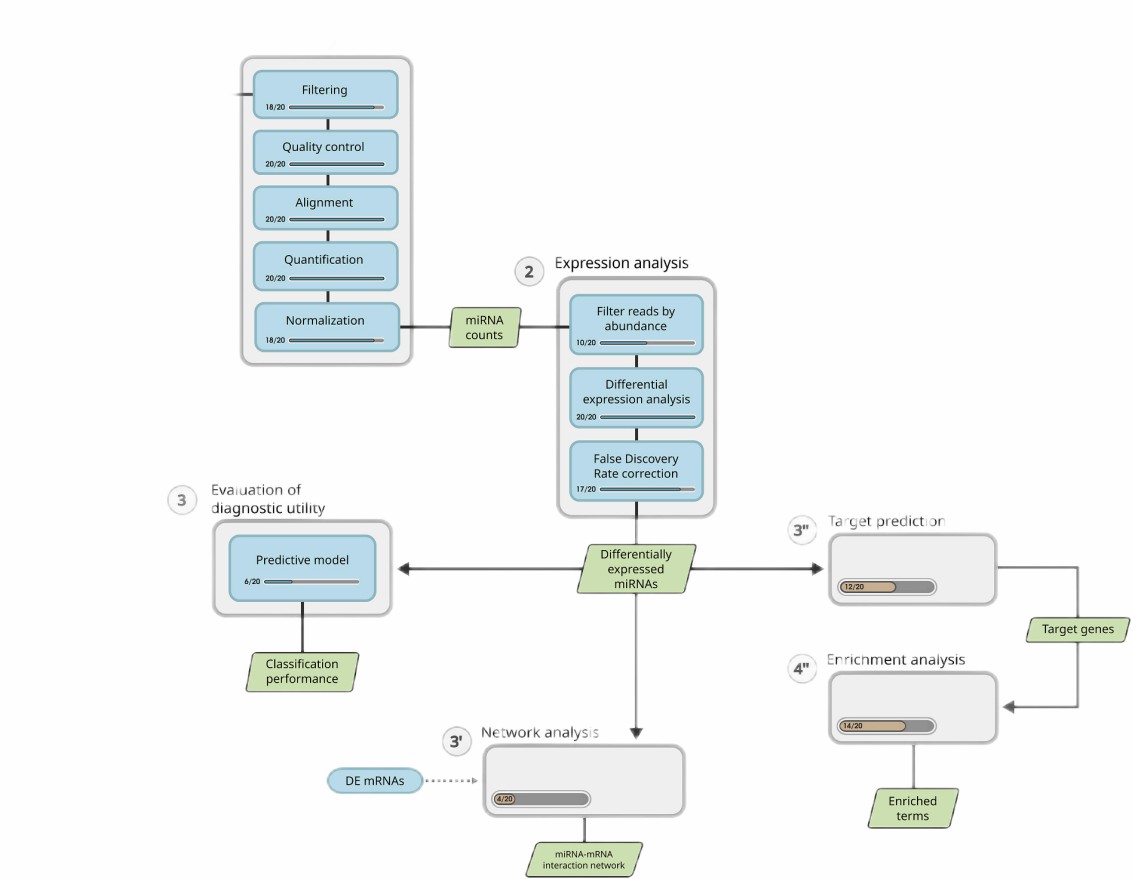 Outline of the general bioinformatics workflow for miRNA-seq
Outline of the general bioinformatics workflow for miRNA-seq
Goals and Objectives of Analysis
The principal objective of microRNA sequencing analysis is to glean an understanding of the intricate networks regulated by miRNAs, and the subsequent influence exerted on fluctuations in gene expressions. Through an in-depth examination of miRNA expression patterns, scientists are positioned to meet several key goals:
i) Decoding miRNA Expression Patterns: miRNA sequencing analysis proffers a robust method for comprehensive probing of miRNA expression patterns amidst diverse biological samples and conditions. By accurately computing miRNA expression quantities, scientists can discern miRNAs that exhibit varying degrees of expression in response to myriad stimuli or aberrancies in health conditions.
ii) Isolating Biomarkers: The comparative investigation of differential expression is instrumental in the isolation of likely miRNAs functioning as biomarkers pivotal for disease diagnosis, prognosis, and responsiveness to treatment. Establishing correlations between miRNA expression profiles and clinical results can unearth potential biomarkers valuable for diagnostic or prognostic applications.
iii) Unveiling New Regulatory Paradigms: The process of miRNA sequencing analysis expedites the unveiling of hitherto unidentified miRNAs and their regulatory correlates - a revelation that illuminates uncharted regulatory mechanisms at play in cellular functions and the manifestation of diseases. By marrying miRNA expression data with mRNA expression profiles, scientists can illuminate gene regulatory networks orchestrated by miRNAs.
Considerations for Pipeline Design
Flexibility for Tailored Outputs: An optimal miRNA sequencing analysis framework should offer flexibility and customization capabilities to meet the varying requirements of different experimental setups, sample types, and research objectives. A modular approach allows for the inclusion of useful additional analysis procedures or required modifications that are highly specific to individual research demands.
Emphasis on Reproducibility and Thorough Documentation: The importance of achieving reproducibility cannot be overstated in the context of pipeline design. Comprehensive documentation of the sequence of analysis, the operational parameters involved, and the versions of software utilized encourages transparency, fostering the reproducibility of results. Such documentation must incorporate scripts, workflows, and standard protocols to seamlessly guide users throughout the analytic process.
Integration of Rigorous Quality Control: It is essential that comprehensive quality control measures are integrated at each step of the pipeline to supervise data quality and pinpoint possible sources of variance. Automated checks for quality control ensure the swift detection and correction of potential issues, thereby bolstering the reliability of the analytic results.
Benchmarking and Validation: It's of paramount importance to validate the performance of the pipeline with a keen eye on its accuracy, sensitivity, and specificity. By benchmarking against established standard datasets or experimental validations, the pipeline's capacity to correctly quantify miRNA expression and pinpoint biologically significant information is validated.
Optimizing Scalability and Efficiency: A well-designed pipeline should be scalable, capable of efficiently handling extensive sequencing datasets. An emphasis on parallelization and optimised utilization of computational resources greatly improves the efficacy of the pipeline, facilitating the swift analysis of high-throughput sequencing data.
Leveraging Illumina for miRNA Sequencing
As a tool for miRNA sequencing, Illumina sequencing technology has come to the fore as a potent platform, endowed with several attributes that render it apt for extensive profiling and analysis of miRNAs. With the application of Illumina sequencing technologies, researchers can delve into the patterns of miRNA expression, unravel regulatory mechanisms, and pinpoint disease correlations with heightened sensitivity and accuracy.
Advantages of Illumina Sequencing Technology
| Aspect | Description |
|---|---|
| Superior Throughput Capacity | Illumina's sequencing platforms, including the NovaSeq and HiSeq series, possess outstanding throughput capabilities, permitting the simultaneous sequencing of thousands to millions of miRNA molecules in one cycle. This scalability leads to cost-effective and efficient miRNA expression profiling across a variety of sample groups and experimental settings. |
| Unmatched Accuracy | Illumina's sequencing technology is universally recognized for its exceptional accuracy, featuring high base-calling precision and minimized error rates. This heightened accuracy is specifically fundamental for miRNA sequencing where exact quantification of miRNA expression and identification of low-abundance miRNAs are critical to discern subtle shifts in gene regulation and biomarker identification. |
| Expanded Dynamic Range | Illumina's sequencing platforms showcase an extended dynamic range, covering several degrees of magnitude in miRNA expression levels. This dynamic range facilitates the detection of both plentiful and scarce miRNAs within the same sequencing procedure, delivering comprehensive coverage of the miRNA landscape and promoting the discovery of novel regulatory components. |
| Adaptable Read Lengths | Illumina sequencing platforms provide the flexibility of read lengths, enabling researchers to customize sequencing procedures to record miRNAs of diverse lengths. This adaptability is especially beneficial for miRNA sequencing, as mature miRNAs typically range between 21 and 24 nucleotides in length. By optimizing read lengths, researchers can accurately capture and quantify miRNA isoforms and variations, enhancing the resolution of miRNA profiling investigations. |
| Optimized Workflow | The workflows of Illumina sequencing are distinguished by their simplicity, efficiency, and the potential for automation. From library preparation to data interpretation, Illumina's optimized workflow minimizes hands-on involvement and maximizes throughput, facilitating researchers in rapidly and consistently processing considerable volumes of samples. This enhanced workflow structure is particularly suited for miRNA sequencing research, where sample throughput and processing effectiveness remain paramount. |
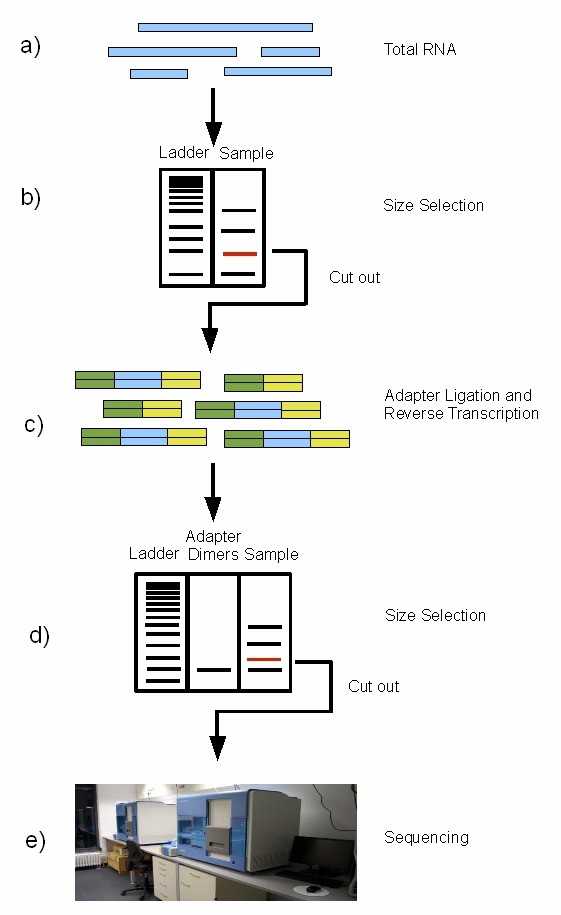 miRNA sequencing procedure on the Illumina
miRNA sequencing procedure on the Illumina
Accessing the MicroRNA Sequencing Database
miRNA sequencing data repositories are cornerstone infrastructures that galvanize data dissemination, foster scientific collaboration, and serve as a platform for integrated, meta-analytical research within the miRNA scientific community. These repositories supply investigators with a portal through which they can access meticulously curated miRNA sequencing data, sourced from a myriad of biological specimens, experimental paradigms, and pathophysiological conditions. As a consequence, they confer opportunities for panoramic investigations into the landscape of miRNA expression profiles and the delineation of their complex regulatory networks.
Importance of Accessing miRNA Sequencing Databases
Data Exchange and Interdisciplinary Productivity: miRNA sequencing databases function as unified platforms that facilitate the global exchange and publication of miRNA sequencing data, originating from a multitude of research entities. By tapping into these collective resources, individual researchers not only contribute their unique datasets, but also can access a plethora of publicly accessible data. This consequently supports cooperative ventures with other investigators, fostering the execution of significant, comprehensive meta-analyses and fusion studies.
Tool for Comparative Examination: miRNA sequencing databases equip researchers with an entryway to a profusion of meticulously curated miRNA expression data, derived from diverse biological contexts, species, and experimental conditions. This acts as a catalyst for comparative investigation into the manifestation patterns of miRNA, pinpointing both conserved and distinct species-specific miRNAs, and facilitates the exploration of miRNA-influenced regulatory networks in relation to both health and disease.
Evidentiary Validation and Consistent Reliability: Utilizing miRNA sequencing databases allows researchers to solidify the validity of their personal findings while simultaneously ensuring the reproducibility of recurrent results through the juxtaposition of their experimental data with universally accessible datasets. Such practices bolster transparency, meticulousness, and fortitude in miRNA inquiry and aid in isolating recurrent miRNA signatures and regulatory routes spanning independent studies.
Formulation of Hypotheses and Novel Discovery: Serving as treasury-like resources, miRNA sequencing databases foster not only the creation of hypotheses but also the revelation of new discoveries by granting researchers the opportunity to delve into unknown miRNA entities, regulatory interactions, and functional associations. Capitalizing on pre-existing datasets enables researchers to unearth groundbreaking miRNA biomarkers, therapeutic targets, and mechanistic knowledge deep-seated within complex biological operations.
Key Resources for Accessing miRNA Sequencing Databases
| Resource | Description |
|---|---|
| miRBase | Serving as a comprehensive reservoir, miRBase encapsulates miRNA sequences, annotations, along with expression data extrapolated from an array of species. It equips researchers with curated information on miRNA sequences, precursor structures, target predictions, and individual expression profiles. This wealth of data facilitates a comparative analysis and further enables functional characterization of miRNAs. |
| Gene Expression Omnibus (GEO) | The Gene Expression Omnibus, a public repository managed by the National Center for Biotechnology Information (NCBI), is instrumental for an efficient archival and distribution of high-throughput sequencing data, including large scale miRNA sequencing datasets. Researchers looking for pertinent miRNA sequencing studies can resort to GEO, and use the raw data available for in-depth analysis. |
| The Cancer Genome Atlas (TCGA) | TCGA leaves no stone unturned in serving as an exhaustive resource, providing a multi-dimensional genomic data for a spectrum of cancer types. This includes miRNA expression profiles, affording researchers with a platform to investigate miRNA dysregulation in cancer. Moreover, it allows for an identification of diagnostic and prognostic biomarkers, and further discovery of therapeutic targets. |
| European Nucleotide Archive (ENA) | Regarded as a principal repository for nucleotide sequence data, ENA houses data derived from research laboratories strewn across the globe. It provides an array of sequencing datasets, notably, the miRNA sequencing data, thus allowing researchers to perform queries, download and systematically analyze data for their individual research motives. |
| Small RNA Expression Atlas | Researchers today can utilize the Small RNA Expression Atlas, a meticulously curated database, to access miRNA expression profiles across various tissues, developmental stages, and experimental conditions. The provision of interactive visualization tools, coupled with data mining capabilities and downloadable datasets, make way for an exhaustive exploration of miRNA expression patterns. |
Evaluating miRNA Sequencing Depth
Evaluating miRNA Sequencing Depth
miRNA sequencing depth refers to the number of sequencing reads generated for miRNA profiling in a given sample. Assessing sequencing depth is crucial in miRNA sequencing experiments as it directly impacts the accuracy, sensitivity, and reliability of miRNA expression quantification.
Importance of Sequencing Depth
Sensitivity of Detection: Ensuring appropriate sequencing depth is fundamental to the reliable detection of less abundant miRNAs, which can often be instrumental in understanding certain biological processes and disease mechanisms. Failing to achieve sufficient sequencing depth may lead to the undetected presence of rare or low-expressed miRNAs, thereby causing skewed expression profiles and an incomplete representation of the miRNA landscape.
Dynamic Range: When an optimal sequencing depth is achieved, it enables the accurate quantification of miRNA expression spanning a broader dynamic range - from highly abundant to minimally expressed miRNAs. A depth-intensive sequencing coverage ensures a linear detection of miRNA expression levels, thus aiding the identification of subtle variations in expressing levels and enabling differential expression analysis across different experimental conditions.
Statistical Confidence: High sequencing depth amplifies the statistical trustworthiness in miRNA expression measurements by reducing sampling variability and augmenting the dependability of differential expression analysis. Ensuring adequate read coverage enables the robust estimation of miRNA expression variance, confidence intervals, and statistical significance, thereby amplifying the reproducibility and interpretability of experimental outcomes.
Discovery Potential: Deep sequencing depth elevates the potential for discovery in miRNA sequencing experiments by facilitating a comprehensive probe into novel miRNAs, isomiRs, and alternative splice variants. A heightened sequencing coverage increases the likelihood of detecting rare or novel miRNA entities, thus fuelling the discovery of novel biomarkers, therapeutic targets, and regulatory elements.
Factors Affecting Sequencing Depth
Sample Complexity: The complexity of a biological sample, inclusive of the variety and abundance of miRNAs, directly influences the required sequencing depth. Samples characterised by a high miRNA diversity, or low miRNA concentration, may command a deeper sequencing coverage to sufficiently capture rare or low-expressed miRNAs.
Library Preparation Efficiency: Differences in the efficiency of library preparation, which incorporates RNA extraction, miRNA enrichment, adapter ligation, and amplification stages, can affect the sequencing depth. Less than optimal library preparation might cause loss in the sample, bias, or ineffective sequencing, leading to reduced sequencing depth and compromised quality of the data.
Sequencing Platform and Chemistry: The selection of sequencing platform and chemistry can sway sequencing depth by virtue of differences in sequencing output, read length, error rate, and sequencing cost. Next-generation sequencing platforms such as Illumina, Ion Torrent, and Oxford Nanopore offer a range of sequencing depths and throughput capacities, which should be considered in the planning of miRNA sequencing experiments.
Experimental Design and Goal: The experimental design, research objectives, and proposed downstream analyses shape the required sequencing depth. Investigations targeting comprehensive miRNA profiling, biomarker discovery, or differential expression analysis, may necessitate a greater sequencing depth as opposed to exploratory or pilot studies.
Strategies for Optimization
Performance of Preliminary Investigations and Pooling of Samples: Preliminary studies and the pooling of samples can assist in predicting the necessary magnitude of sequencing depth, considering the complexity and variability inherent to experimental samples. Initiation of pilot sequencing projects paves the way for researchers to fine-tune library preparation protocols, assess the expected sequencing output, and determine the optimal depth for future large-scale experimentation.
Standardization and Quality Assurance: The incorporation of standardization techniques and the reinforcement of strict quality control measures can augment uniformity in sequencing depth and enhance the reliability of data. Standardization methods such as the Reads Per Million (RPM) or Transcripts Per Million (TPM) adjust for variations in library size and depth of sequencing, thereby enabling accurate comparisons of miRNA expression levels across multiple samples.
Analysis Adjusted for Depth: Modifying the analysis pipeline to account for the variability in sequencing depth can amplify the robustness and comparability of miRNA expression profiles. Depth-adjusted normalization strategies, such as DESeq2 or edgeR, incorporate sequencing depth as a vital covariate in differential expression analysis, mitigating the biases born from variations in sequencing depth.
Striking a Balance between Cost and Coverage: Balancing the depth of sequencing with practical cost considerations is crucial to optimize experimental results and maximize the utilization of resources. Researchers should evaluate the trade-offs between the depth of sequencing, the quality of data, and budgetary constraints.
Future Perspectives in miRNA Sequencing Research
As the technology for miRNA sequencing continues to evolve, we can envisage several future directions that promise to enhance not only our grasp of miRNA biology, but also its practical clinical applications:
Single-Cell miRNA Sequencing: With impressive strides being made in the field of single-cell sequencing technologies, one can anticipate unprecedented opportunities surfacing to decipher cell-specific miRNA expression profiles and associated regulatory networks. This, in turn, can demystify the heterogeneity of cells and their reactionary dynamics within the contexts of both health and pathology.
Long-Read Sequencing: By combining long-read sequencing platforms, such as Oxford Nanopore and PacBio, with miRNA sequencing, the potential exists for more thorough characterizations of miRNA isoforms, alternative splicing events, and structural variations. This integration effectively elevates the precision and resolution of miRNA profiling.
Integrative Multi-Omics Analysis: The fusion of miRNA sequencing data with disparate omics data (including, but not limited to mRNA sequencing, epigenomics, and proteomics) promises to enable a more multi-dimensional exploration into gene regulatory networks. This approach can unlock deeply woven interactions and functional correlations underlying a multitude of intricate biological processes.
Clinical Translation and Biomarker Discovery: As concerted efforts to facilitate clinical corroboration as well as biomarker discovery utilizing miRNA sequencing continue unabated, the prospect of discerning innovative diagnostic, prognostic, and therapeutic targets in an assortment of diseases comes into sharper focus. This, in a broader sense, lay a solid groundwork for subsequent developments in personalized medicine and precision oncology interventions.
In summary, miRNA sequencing represents a powerful tool for deciphering the intricate landscape of miRNA-mediated gene regulation, offering insights into fundamental biological processes and disease mechanisms. By leveraging advancements in sequencing technologies, bioinformatics analysis, and integrative approaches, researchers can unravel the complexities of the miRNA and harness its potential for transformative applications in biomedical research and clinical practice.
References:
- Haseeb, A., Makki, M.S., Khan, N.M. et al. Deep sequencing and analyses of miRNAs, isomiRs and miRNA induced silencing complex (miRISC)-associated miRNome in primary human chondrocytes. Sci Rep 7, 15178 (2017).
- Jardillier R, Koca D, Chatelain F, Guyon L. Optimal microRNA Sequencing Depth to Predict Cancer Patient Survival with Random Forest and Cox Models. Genes (Basel). 2022
- Campbell JD, Liu G, Luo L, Xiao J, Gerrein J, Juan-Guardela B, Tedrow J, Alekseyev YO, Yang IV, Correll M, Geraci M, Quackenbush J, Sciurba F, Schwartz DA, Kaminski N, Johnson WE, Monti S, Spira A, Beane J, Lenburg ME. Assessment of microRNA differential expression and detection in multiplexed small RNA sequencing data. RNA. 2015
- Khamina K, Diendorfer AB, Skalicky S, Weigl M, Pultar M, Krammer TL, Fournier CA, Schofield AL, Otto C, Smith AT, Buchtele N, Schoergenhofer C, Jilma B, Frank BJH, Hofstaetter JG, Grillari R, Grillari J, Ruprecht K, Goldring CE, Rehrauer H, Glaab WE, Hackl M. A MicroRNA Next-Generation-Sequencing Discovery Assay (miND) for Genome-Scale Analysis and Absolute Quantitation of Circulating MicroRNA Biomarkers. Int J Mol Sci. 2022
- Potla P, Ali SA, Kapoor M. A bioinformatics approach to microRNA-sequencing analysis. Osteoarthr Cartil Open. 2020 Dec 19;3(1):100131. doi: 10.1016/j.ocarto.2020
- Pérez-Rodríguez D, López-Fernández H, Agís-Balboa RC. Application of miRNA-seq in neuropsychiatry: A methodological perspective. Comput Biol Med. 2021 Aug;135:104603. doi: 10.1016/j.compbiomed.2021
- Oh JN, Son D, Choi KH, Hwang JY, Lee DK, Kim SH, Lee M, Jeong J, Choe GC, Lee CK. MicroRNA expression data of pluripotent and somatic cells and identification of cell type-specific MicroRNAs in pigs. Data Brief. 2020
- Motameny S, Wolters S, Nürnberg P, Schumacher B. Next Generation Sequencing of miRNAs - Strategies, Resources and Methods. Genes (Basel). 2010


 Sample Submission Guidelines
Sample Submission Guidelines
