How to Detect Microbial Sulfur Cycling Networks by Metagenomes?
What is Microbial Sulfur Cycling?
Sulfur, an indispensable nutrient for all life forms, serves as a vital building block for amino acids such as methionine and cysteine. Abundant sulfur reservoirs exist in nature, particularly in the marine ecosystem, where sulfate and sulfide are stored in copious amounts. The transformation of sulfur compounds with varying valence states primarily relies on microorganisms, each equipped with distinct metabolic functions, contributing to the intricate sulfur cycle. Under diverse conditions, sulfur-oxidizing bacteria facilitate the conversion of low-valence sulfur components into sulfate, whether in aerobic or anaerobic settings. Conversely, sulfate-reducing bacteria, active under anaerobic conditions, reduce sulfate to sulfide, completing this essential biological cycle.
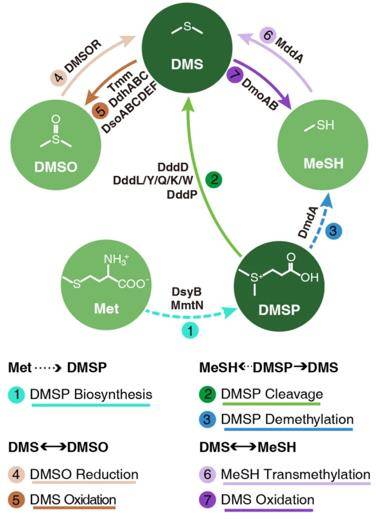 The conceptual sketch of known key proteins and pathways involved in microbial DMS/DMSP cycling. (Teng et al., 2021)
The conceptual sketch of known key proteins and pathways involved in microbial DMS/DMSP cycling. (Teng et al., 2021)
Metagenomic Data Mining
- Quantification and Comparative Analysis of Sulfur Cycle Functional Genes: Detailed gene abundance calculation of sulfur cycle functional genes.
- Sulfur Cycle Functional Gene Pathway Map: Visualization of key pathways and their interactions.
- Comparative Analysis of Beta Diversity Between Samples/Groups: Uncover the diversity in sulfur cycle genes across different samples or groups.
- Host Microbial Identification and Joint Analysis with Functional Genes: Linking microbial communities to the functional genes they carry, providing a holistic perspective.
Please read our article Introduction to Shotgun Metagenomics, from Sampling to Data Analysis.
Process of Research
- KEGG Annotation Results from Metagenome Standard Analysis: Utilizing KEGG annotation to identify relevant genes in the sulfur cycle.
- Integration of Carbon, Nitrogen, Sulfur, and Phosphorus Cycling Pathways: Comprehensive understanding of the interconnected metabolic cycles.
- Detailed Gene Abundance Calculation: Precise quantification of gene abundance for each pathway (carbon, nitrogen, sulfur, and phosphorus) in every sample.
- Inter-Sample Gene Abundance Comparisons: Unveiling the variations and relationships among different samples.
Case Study: Biogeographic Characterization of the Dimethylsulfide and Dimethylmercaptopropionate Cycling
Dimethylmercaptopropionate (DMSP) is a common marine compound vital to bacterial nutrition and the global sulfur cycle. Microorganisms metabolize DMSP into volatile dimethyl sulfide (DMS), a "climate-active" gas with significant implications for atmospheric chemistry and global climate change.
In this study, they conducted metagenomic and metatranscriptomic analyses on seawater samples from polar and low-latitude oceans, shedding light on the biogeographic traits of microbial genes involved in the DMS/DMSP cycle.
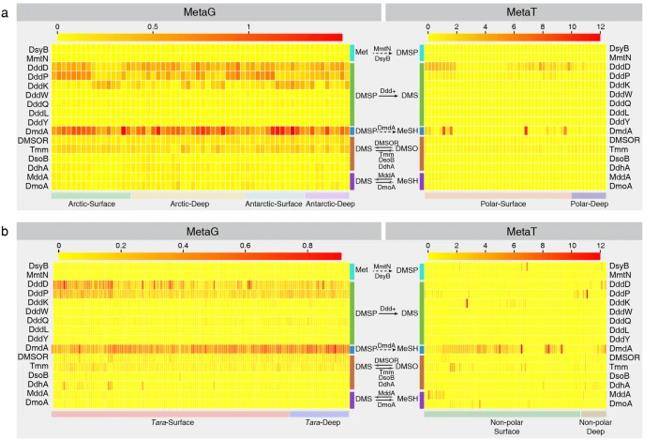 Relative abundance of potential genes involved in bacterial DMS/DMSP cycling. (Teng et al., 2021)
Relative abundance of potential genes involved in bacterial DMS/DMSP cycling. (Teng et al., 2021)
Their findings reveal that polar oceans exhibit higher gene abundance related to the DMS/DMSP cycle compared to low-latitude oceans, demonstrating a distinct biogeographic distribution. Key genes, such as DMSP demethylase (DmdA), DMSP cleaving enzymes (DddD, DddP, and DddK), and trimethylamine monooxygenase (Tmm), which oxidizes DMS to dimethylsulfoxide, are central players in the global DMS/DMSP cycle.
Within the polar marine DMS/DMSP cycle, α-amoeba and γ-amoeba have crucial roles. Surprisingly, the biogeographic pattern of the DMS/DMSP cycle in polar oceans is primarily associated with sampling depth, rather than geographic distance, indicating that the distribution is not constrained by dispersal limitations but rather influenced by variations in surface and deep-water habitats.
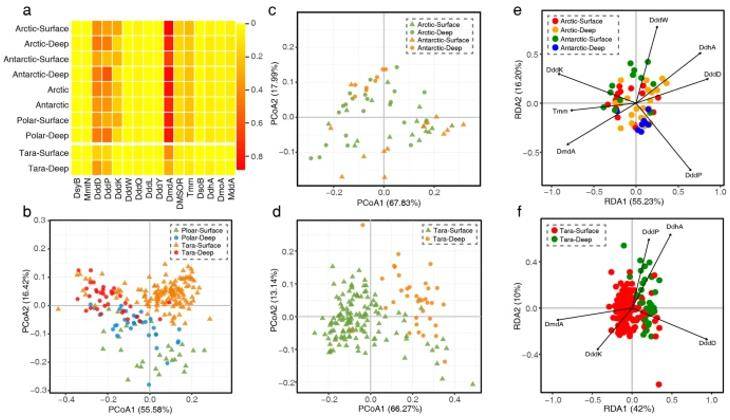 Analyses of inter-sample similarity among the polar and non-polar seawater samples. (Teng et al., 2021)
Analyses of inter-sample similarity among the polar and non-polar seawater samples. (Teng et al., 2021)
In conclusion, this research underscores the significance of microbial sulfur cycling in marine ecosystems, with the utilization of metagenomic data providing invaluable insights into the distribution and diversity of key genes involved in the DMS/DMSP cycle, ultimately contributing to our understanding of the Earth's biogeochemical processes.
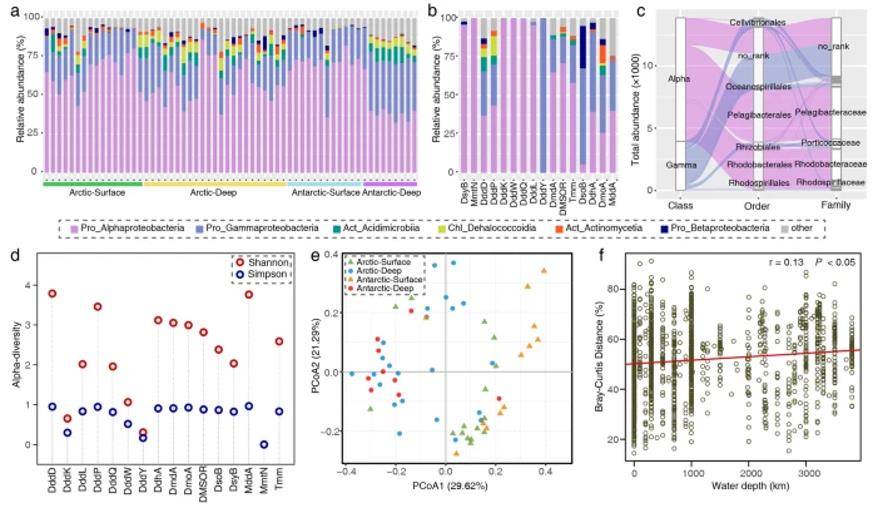 Phylogenetic diversity of DMS/DMSP cycling-related genes in the Arctic and Antarctic oceans. (Teng et al., 2021)
Phylogenetic diversity of DMS/DMSP cycling-related genes in the Arctic and Antarctic oceans. (Teng et al., 2021)
Reference:
- Teng, Zhao-Jie, et al. "Biogeographic traits of dimethyl sulfide and dimethylsulfoniopropionate cycling in polar oceans." Microbiome 9.1 (2021): 1-17.


 Sample Submission Guidelines
Sample Submission Guidelines
