Exploring the Methane Cycling through Metagenomic Sequencing
Methane Cycling
Methane (CH4), is a ubiquitous compound found in nature. It stands as the most basic organic substance and the simplest hydrocarbon. However, its environmental significance cannot be understated. Methane ranks as a potent greenhouse gas, second only to carbon dioxide, exerting a substantial influence on our planet's ecosystem and global climate dynamics. It possesses a global warming potential 28 times greater than that of CO2, contributing to approximately 20% of the overall global warming effect.
The methane metabolic processes currently at the forefront of scientific investigation are methanogenesis and methane oxidation. Within these processes, microorganisms assume pivotal roles. More than two-thirds of global methane emissions originate from microbial sources, with a significant portion being attributed to methane-producing microorganisms, known as methanogens. These microorganisms predominantly thrive in anaerobic, oxygen-deprived environments.
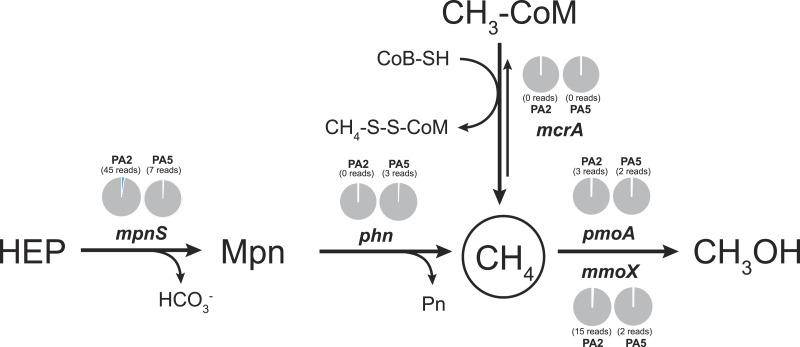 Methane cycling potential in the Arabian Sea oxygen minimum zone.
Methane cycling potential in the Arabian Sea oxygen minimum zone.
Metagenomic Sequencing and Methane Cycle Gene Analysis
The field of Metagenomic Methane Cycle Gene Analysis employs metagenomic sequencing techniques to unravel the functional capacities and activities of microorganisms within diverse habitats. This approach serves to reconstruct their contributions to the intricate methane cycle. By utilizing advanced sequencing technologies, microbial genome sequences are obtained from various ecosystems. Subsequently, these metagenomic datasets are annotated to reveal their involvement in methane metabolism functions and to identify the species involved. This process is facilitated through the Methane Cycle Metabolism Database.
The database is instrumental in annotating metagenomic sequencing data from a wide array of environments, including hot spring sediments, marine sediments, freshwater ecosystems, peatlands, and tundra regions. The results provide an efficient, specific, and comprehensive analysis of both the structural and functional aspects of microbial communities involved in methane metabolism. This high-precision approach enhances our understanding of microbial-driven methane cycling and its intricate interactions with other elements in the environment.
Case: Nitrogen and Methane Cycles in the Arabian Sea's Oxygen Minimum Zones
In the oxygen minimum zones (OMZs) of the global ocean, where oxygen concentrations plummet to less than one percent, unique microbial communities drive intricate biogeochemical cycles, including those of nitrogen and methane. These regions are essential contributors to oceanic nitrogen loss, playing a crucial role in shaping the chemical composition of the world's oceans. Among OMZs, the Arabian Sea stands out as a notable site where both anammox (anaerobic ammonium oxidation) and denitrification processes have been documented.
To gain a deeper understanding of the genetic potential for nitrogen and methane cycling within the Arabian Sea OMZ, a comprehensive metagenomic study was conducted, focusing on two distinct zones: the upper and core regions of the OMZ.
In the upper zone of the Arabian Sea OMZ, they observed a diverse community of Thaumarchaeota, suggesting their involvement in aerobic ammonium oxidation. Thaumarchaeota are known for their capability to oxidize ammonium aerobically, shedding light on the upper OMZ's nitrogen cycling processes. The presence of this diverse Thaumarchaeota community points to their active participation in this essential step of the nitrogen cycle.
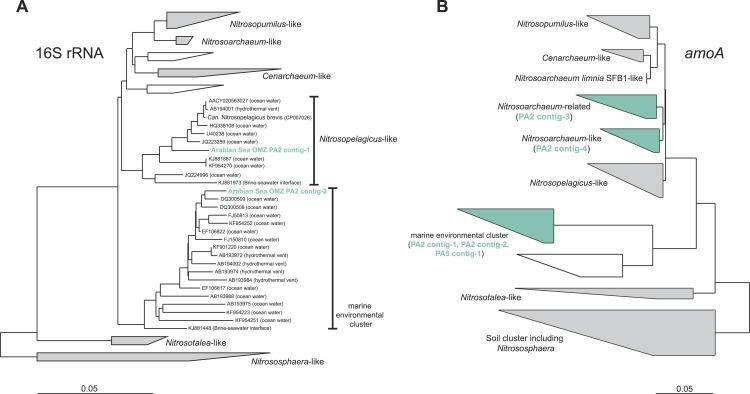 Phylogenetic inference of thaumarchaeal contigs assembled from the OMZ metagenomes. (Lüke et al., 2016)
Phylogenetic inference of thaumarchaeal contigs assembled from the OMZ metagenomes. (Lüke et al., 2016)
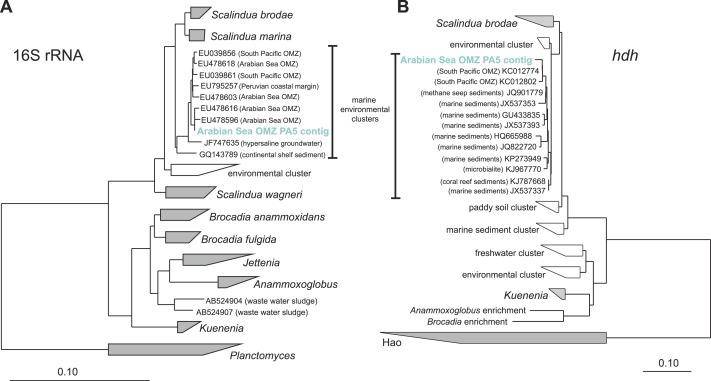 Phylogenetic inference of Scalindua-related contigs assembled from the OMZ metagenomes. (Lüke et al., 2016)
Phylogenetic inference of Scalindua-related contigs assembled from the OMZ metagenomes. (Lüke et al., 2016)
Moving deeper into the core zone of the Arabian Sea OMZ, they observed a low diversity of Scalindua-like anammox bacteria, which played a significant role in nitrogen loss. Anammox bacteria are renowned for their unique ability to convert ammonium and nitrite into nitrogen gas under anaerobic conditions, thus contributing to the reduction of nitrogen compounds in the ocean.
The study also identified Nitrospina spp. and a novel lineage of nitrite-oxidizing organisms as the key players in aerobic nitrite oxidation within the OMZ. Both of these groups were present in roughly equal abundance, emphasizing their shared importance in maintaining nitrogen cycling processes in this environment.
Furthermore, their research indicated the potential for dissimilatory nitrate reduction to ammonia (DNRA) in the OMZ. The responsible microorganisms were not yet identified but were found to harbor a divergent nrfA gene. DNRA is an essential step in the nitrogen cycle, converting nitrate into ammonia and influencing the availability of essential nutrients in the ocean.
While the metagenomic data did not provide conclusive evidence for active methane cycling in the Arabian Sea OMZ, they did detect a low abundance of novel alkane monooxygenase diversity. These findings suggest that methane metabolism may be occurring, albeit at a relatively low level.
In conclusion, their comprehensive metagenomic study of the Arabian Sea OMZ revealed a dynamic and diverse community of microorganisms participating in the nitrogen cycle, encompassing aerobic ammonium oxidation, anammox, aerobic nitrite oxidation, and the potential for DNRA. These findings shed light on the genetic potential for active nitrogen cycling in this unique environment and unveiled hitherto overlooked lineages of bacteria involved in carbon and nitrogen cycling.
Reference:
- Lüke, Claudia, et al. "Metagenomic analysis of nitrogen and methane cycling in the Arabian Sea oxygen minimum zone." PeerJ 4 (2016): e1924.


 Sample Submission Guidelines
Sample Submission Guidelines
