Exosme Research Overview
Extracellular Vesicles (EVs), commonly referred to as exosomes, are a ubiquitous feature in the realm of cellular biology. Research has firmly demonstrated that these vesicles are not only secreted by eukaryotic cells but are also produced by all prokaryotic cells, extending their prevalence. Within multicellular organisms, exosomes are disseminated into a range of bodily fluids, encompassing blood, urine, saliva, breast milk, amniotic fluid, ascites, cerebrospinal fluid, bile, semen, and more.
These vesicles are often referred to by different names in the scientific literature, based on their origin, nature, and characteristics. While there isn't a universally accepted definition, there is a general categorization that distinguishes between exosomes, shedding vesicles, microparticles, and microvesicles (MV), which typically range in size from 150 to 1000 nanometers and are formed through budding from the cell membrane. In contrast, exosomes are a subtype of EVs characterized by their lipid bilayer membrane structure. They have a diameter of approximately 30-100 nanometers and are formed intracellularly within multivesicular bodies (MVBs) through the fusion of multiple endosomes with the cell membrane.
Composition of Exosomes
Exosomes, comprise a diverse array of components. In 1981, Trams and colleagues first conceptualized the idea of exosmes when they discovered vesicles secreted by cells exhibiting extracellular enzyme activity. This concept was later applied to vesicles released during reticulocyte differentiation, which typically have a diameter ranging from approximately 30 to 100 nanometers. Over the subsequent years, it became evident that various cell types, including B lymphocytes, dendritic cells, T cells, mast cells, neuronal cells, endothelial cells, and epithelial cells, can secrete exosmes. Furthermore, multiple body fluids, such as blood, urine, saliva, breast milk, amniotic fluid, ascites, cerebrospinal fluid, bile, and semen, also contain exosmes.
Exosmes share common protein components involved in membrane transport and fusion, including GTPases, annexins, flotillins, as well as transmembrane proteins like CD9, CD63, CD81, and CD82. Heat shock proteins (Hsc70 and Hsp90), vesicle biogenesis proteins (Alix and TSG101), lipid-associated proteins, and phospholipases are also present in all types of exosmes. Currently, over 4500 different proteins have been identified in exosmes. In addition to proteins, exosmes contain a variety of lipids such as cholesterol, sphingolipids, and long-chain glycerophospholipids.
Moreover, exosomes house a diverse array of RNA species, comprising messenger RNA (mRNA), microRNA (miRNA), and various non-coding RNAs, notably small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), small Cajal body-specific RNA (scaRNA), piwi-interacting RNA (piRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA). The majority of these RNA molecules identified within EVs exhibit lengths ranging from 20 to 200 nucleotides. This spectrum includes full-length miRNAs and tRNAs, as well as fragmented forms of mRNA and rRNA.
What are The Functions of Exosomes?
Exosomes, initially perceived as cellular debris upon their discovery, have garnered increased attention in recent years due to their pivotal role in intercellular communication. These diminutive membrane-bound vesicles are now recognized for their content of cell-specific proteins, lipids, and nucleic acids, which function as signaling molecules when transmitted to recipient cells, thus influencing their cellular functions. Exosomes play crucial roles in a wide array of physiological and pathological processes, including immune system antigen presentation, tumor growth and metastasis, tissue regeneration post-injury, programmed cell apoptosis, vascular regeneration, inflammation, blood coagulation, pathogen propagation, and more. Different cell types secrete exosomes with distinct compositions and functions, rendering them valuable as biomarkers for disease diagnosis. Due to their lipid bilayer membrane structure, exosomes provide effective protection for their cargo and can be selectively targeted to specific cells or tissues, making them a promising platform for drug delivery.
Amid the expanding emphasis on precision medicine, there is a growing focus on achieving accurate disease diagnosis and tailored treatments. Exosomes, an emerging and novel realm of investigation, present a promising avenue in this endeavor. Their ubiquity in the body and the ease of their retrieval position them as a potentially effective approach for disease diagnosis and treatment. This underscores their significant potential in advancing the field of precision medicine.
Isolation of Extracellular Vesicles
Ultracentrifugation
Ultracentrifugation is a common method for the isolation of exosmes and can be employed alongside sucrose density gradient centrifugation or sucrose cushion separation for low-abundance exosmes. Although this method is straightforward in operation, it has limitations, such as the capacity to process only up to six samples simultaneously due to rotor constraints, and the yield can be inconsistent, which may be related to the type of rotor used. Ultracentrifugation also requires a substantial amount of starting material, and the yield and purity of exosmes obtained may be questionable. Additionally, repetitive centrifugation steps can potentially damage vesicles, leading to a reduction in their quality. However, when ultracentrifugation is combined with sucrose gradient centrifugation, it can yield high-purity exosmes, although this approach is relatively time-consuming.
Magnetic Bead Immunoaffinity
exosmes possess specific surface markers such as CD63 and CD9 proteins. By incubating exosmes with magnetic beads coated with antibodies against these markers, exosmes can be captured and isolated. Magnetic bead immunoaffinity offers advantages including high specificity, ease of operation, and minimal impact on the integrity of exosme morphology. However, it has drawbacks, including lower efficiency, susceptibility of exosme biological activity to pH and salt concentration variations, which may hinder downstream experiments, and limited widespread applicability.
PEG-Based Precipitation Method
Polyethylene glycol (PEG) can be employed to co-precipitate hydrophobic proteins and lipid molecules. Initially utilized for virus collection from samples like serum, it is now also used for exosome precipitation. The principle behind this technique may involve competitive binding with free water molecules.
However, the utilization of PEG for exosome precipitation presents several challenges. These include issues with purity and yield, an elevated presence of impurities (leading to false positives), non-uniform particle sizes, the formation of intractable aggregates that are challenging to remove, and the potential disruption of exosomes by mechanical forces or chemical additives such as Tween-20. Consequently, studies employing PEG precipitation for exosome isolation may face scrutiny when published.
Reagent-Based Extraction
In recent years, various commercial exosome extraction reagent kits have emerged in the market. Some of these kits employ specially designed filters to remove impurities, while others utilize Size-Exclusion Chromatography (SEC) for separation and purification. There are also kits that rely on compound precipitation methods to precipitate exosomes. These reagent kits do not require specialized equipment, and as they undergo continuous updates and improvements, their extraction efficiency and purification effectiveness gradually increase. Consequently, they are gradually replacing ultracentrifugation methods and gaining wider adoption.
Identification of Isolated Exosomes
Electron Microscopy Identification
The most straightforward approach to identify exosomes involves their observation under an electron microscope. In a typical exosome electron microscopy examination, these vesicles appear as spherical or cup-shaped structures, exhibiting sizes within the range of 30 to 100 nanometers.
Western Blot Identification
Following exosome isolation, Western blot (WB) analysis can be conducted with specific exosome-associated proteins. WB typically includes proteins related to multivesicular bodies (MVBs) synthesis, such as Alix and TSG101, as well as tetraspanin transmembrane proteins (CD9, CD63, CD81, CD82). It is also advisable to include non-exosome markers in the WB, such as GRP94, EEA1, Cytochrome c, GM130, PMP70, Calreticulin, prohibitin, which are absent in exosomes and are exclusive to total cell proteins.
How to Extract Exosomes RNA
Once extracellular vesicles (EVs) have been isolated, RNA can be directly extracted using the following methods:
Isolate EVs using a commercial kit, and then extract RNA using an RNA extraction kit.
Isolate EVs using a commercial kit, and then employ a matching RNA extraction kit designed for EVs.
Our comparative experiments have shown that strategy 1 yields superior extraction results.
Exosome Research Application
In vivo imaging confirms widespread exosome delivery
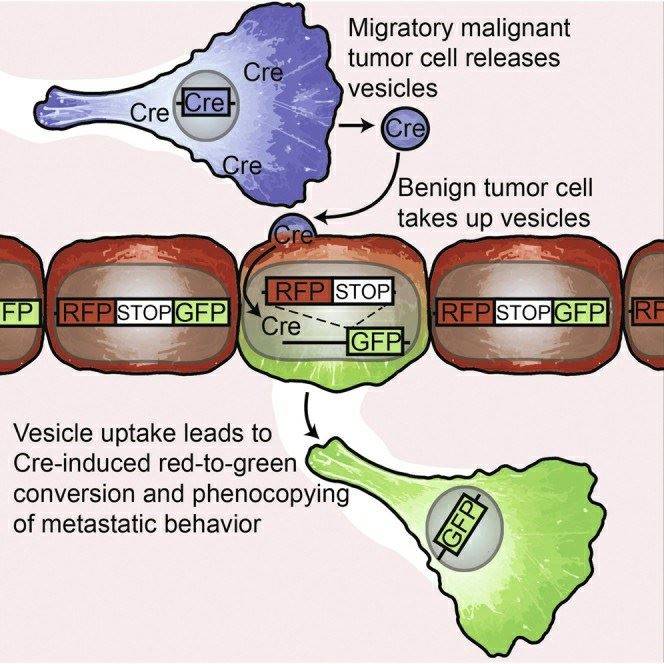
On May 21, 2015, a groundbreaking discovery in the field of cancer metastasis research was published in the prestigious journal "Cell." The study was conducted by a team of researchers from the Hubrecht Institute. Their research confirmed that cancer cells in the process of metastasis can impart specific behaviors to less malignant cancer cells. This significant finding offers valuable insights into the behavior of cancer and holds promise for improving cancer diagnosis and treatment.
Cancer arises from the accumulation of genetic errors in cell DNA. Some of these errors cause cells to disregard signals that control growth and differentiation, leading to uncontrolled cell division. This ultimately results in tumor growth, with tumor cells accumulating more and more errors. Due to the fact that each of these DNA errors in individual tumor cells is unique (tumor heterogeneity), the behavior of these individual tumor cells can also differ. This complexity makes cancer challenging to treat, as specific combinations of DNA errors can render some tumor cells resistant to treatment.
To gain a better understanding of the behavior of individual tumor cells, researchers developed specialized microscopy techniques, including in vivo microscopy, to capture the actions of cancer cells within living organisms. They assigned different colors to tumor cells carrying distinct DNA errors by using fluorescent markers. By imaging these colored cells, it became evident which cells were mobile and capable of initiating metastasis within the body.
Invasive malignant cancer cells (blue) release very small extracellular vesicles (EVs), such as exosomes and microvesicles, containing molecules that can induce cellular malignancy. Less malignant cells (red) take up these EVs, altering their behavior. Less malignant cells become more malignant and initiate metastasis (green).
Scientists have also confirmed that these perilous EVs can travel through the bloodstream to other tumor sites, replicating malignant behavior over long distances.
Exosomes determine organ specificity of tumor metastasis
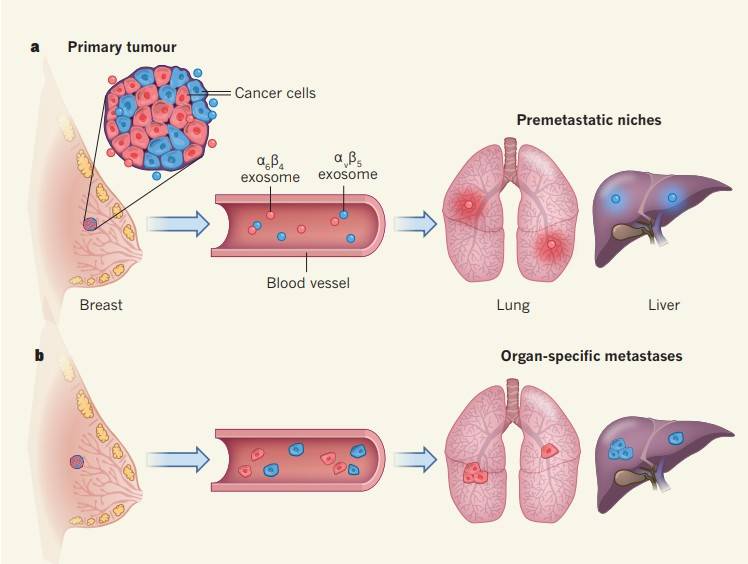
The leading contributor to cancer-related deaths is the dissemination of cancer cells from their primary site to remote organs via the circulatory system. This metastatic process is not stochastic in nature. Rather, specific subsets of cancer cells follow intricate molecular pathways that enable them to selectively target particular organs and establish colonies within them. This directed migration involves a dynamic interplay between the cancer cells, often colloquially referred to as the "seeds," and the microenvironment within the destination organ, commonly known as the "soil."
Recent scientific investigations have unveiled an intriguing phenomenon suggesting that, before the arrival of metastatic "seeds," they have the potential to modulate the microenvironment, often referred to as the "soil," through the utilization of extracellular vesicles, specifically, exosomes. Exosomes are succinctly characterized as minute extracellular vesicles with the ability to transport an array of molecules, encompassing proteins, lipids, and nucleic acids, between cells. These vesicles possess the unique capability to navigate the circulatory system, enabling them to reach distant anatomical sites. This intricate interplay orchestrates a molecular dialogue, ultimately preparing the terrain for the ensuing tumor metastasis.
When cancer cell-derived exosomes are injected into mice, these exosomes tend to accumulate in the organs where cancer cells are inclined to metastasize. Furthermore, these organ-specific exosomes can interact with different cell types. For example, exosomes targeting the lungs adhere to endothelial cells within the lung, while those targeting the liver enter Kupffer immune cells. Injecting cancer cell-derived exosomes into the same cell line confirms that exosomes facilitate organ-specific tumor metastasis.
Researchers have also made an intriguing discovery—exosomes from breast cancer cells that typically metastasize to the lungs can redirect another type of tumor cell, which usually migrates to the bones, to metastasize to the lungs. This finding further underscores that the characteristics of tumor cell metastasis are not autonomous but are influenced by external factors.
The researchers provided several clues about how exosomes influence organ-specific metastasis. They found that exosomes targeting different organs possess distinct cell adhesion receptor proteins, specifically integrins on their surface. Different types of exosomes show a preference for entering organs that have a substantial number of ligands corresponding to their surface integrins. For instance, αVβ5 integrin directs exosomes to the liver, while α6β4 integrin directs them to the lungs. Moreover, inhibiting the expression of exosomes or integrins can inhibit cancer metastasis. Finally, when exosomes invade the target organ, they trigger the synthesis of S100 proteins, promoting inflammation and cell migration, and activate Src proteins—all of which lay the foundation for cancer cell metastasis.
Presence of Circular RNA in Exosomes
Circular RNA (circRNA) is a ubiquitous and exceptionally stable RNA molecule with substantial regulatory significance in mammalian gene expression. Concurrently, exosomes, minute membrane vesicles derived from endocytic processes, have emerged as a subject of considerable research attention. In a groundbreaking investigation recently conducted by the research team led by Shenglin Huang and Xianghuo He at Fudan University's Institute of Oncology, an intriguing revelation came to light. This study unveiled a wealth of circRNAs within exosomes, a previously unrecognized phenomenon. The notable findings from this research endeavor were published in the journal Cell Research.
The research team extracted total RNA (excluding ribosomal RNA) from both MHCC-LM3 liver cancer cells and exosomes derived from these cells. They conducted RNA-seq analysis and found that the circRNAs in exosomes are much more abundant than those in the source cells.
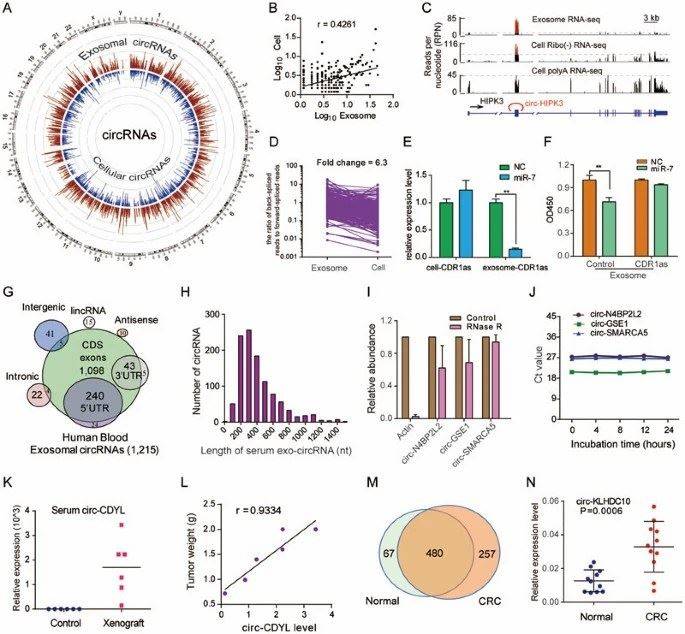 A: Circos diagram displays circRNA in cells and cell-derived exosomes; D: The ratio of circRNA to linear RNA is six times higher in exosomes than in cells. E: After transfection of HEK293T cells with miR-7 mimics, the levels of CDR1as in both the cells and cell-derived exosomes were assessed.
A: Circos diagram displays circRNA in cells and cell-derived exosomes; D: The ratio of circRNA to linear RNA is six times higher in exosomes than in cells. E: After transfection of HEK293T cells with miR-7 mimics, the levels of CDR1as in both the cells and cell-derived exosomes were assessed.
Researchers refer to the circRNA present in exosomes as exo-circRNA. Exo-circRNA is over two times more abundant than the circRNA found in the source cells. Furthermore, the circRNA entering exosomes surpasses the quantity of linear RNA.
Prior research has previously demonstrated the capacity of circular RNAs (circRNAs) to interact with miRNAs. Given the abundance of miRNAs within exosomes, investigators delved deeper into the intricate relationship between circRNAs and miRNAs. Specifically, they focused on CDR1as, acknowledged as a sponge for miR-7. To explore this relationship, miR-7 mimics were transfected into HEK293T cells, and the CDR1as levels were evaluated in both the cells and their exosomes. Notably, the study revealed a substantial reduction in CDR1as levels within exosomes upon miR-7 transfection, whereas the decline in the cells was marginal. This finding implies that the packaging of circRNAs into exosomes is a regulated process, influenced, at least in part, by the modulation of relevant miRNA levels within the source cells.
In addition to cell-derived exosomes, researchers also discovered a significant amount of intact and stable circRNA in human serum exosomes. Furthermore, the circRNA in serum exosomes from colorectal cancer patients showed noticeable differences compared to those from healthy individuals. Exo-circRNA originating from tumors holds promise as a potential biomarker for cancer detection.
Exosome Transcriptome Sequencing
Exosomes primarily contain RNA, including non-coding RNA (small RNA, lncRNA, circRNA) and coding RNA (mRNA). They play a crucial role in intercellular communication and gene expression regulation. Given their advantages, such as amplifiability and high representation, RNA in exosomes holds a dominant position in the research of exosome content and functional exploration. Additionally, small RNA consists of various types of small RNAs, including miRNA, piRNA, tsRNA, YRNA, each with distinct functions.
Distinctions are evident between exosomal RNA and RNA extracted from standard tissue samples, prompting investigations into the composition of exosomal RNA. CD Genomics has pioneered a comprehensive workflow that is grounded in authentic samples, encompassing the isolation of exosomes, the extraction of exosomal RNA, the construction of transcriptome libraries, the establishment of miRNA libraries, and the subsequent analysis of the acquired data.
References:
- In Vivo imaging reveals extracellular vesicle-mediated phenocopyingof metastatic behavior. Cell. 2015.
- Cancer: Organ-seeking vesicles. Nature. 2015.
- Circular RNA is enrichedand stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015.


 Sample Submission Guidelines
Sample Submission Guidelines
