Deep Sequencing
What Is Deep Sequencing?
Deep sequencing entails the meticulous analysis of genomic regions through repeated sequencing, often spanning hundreds or even thousands of iterations. This cutting-edge next-generation sequencing (NGS) approach empowers researchers to uncover exceedingly rare clone types, cells, or microbial entities, even when they comprise as little as 1% of the original sample.
Employing both short and long read lengths, deep sequencing enables the detection of elusive cell populations crucial in cancer and microbial research, among other fields. Its utility extends to discerning minute variations within tumors, a task complicated by the common occurrence of normal cell contamination in cancer samples and the presence of diverse cancer cell subclones within the tumor itself.
CD Genomics short-read sequencing and long-read sequencing platforms facilitate the robust analysis of DNA/genomes. This advanced sequencing approach allows for comprehensive and efficient examination of genetic material, providing valuable insights into the molecular landscape and potential biomarkers associated with various conditions.
What Is Sequencing Depth?
Following the advent of deep sequencing following PCR and first-generation sequencing, genetic testing has transitioned into a phase marked by the widespread adoption of NGS. Within this landscape, sequencing depth emerges as a pivotal metric, signifying the specificity and sensitivity of deep sequencing. But what precisely is sequencing depth? In professional terms, sequencing depth denotes the ratio of total bases (bp) covering the target region to the size of the target genome. Put simply, it represents the frequency with which the bases of the target gene are iteratively interpreted. Analogously, the iterative interpretation of bases during deep sequencing can be likened to honing a repetitive task, where increased repetition correlates with greater skillfulness. Consequently, inadequate sequencing depth significantly compromises accuracy.
What Is Sequencing Coverage?
Sequencing coverage, on the other hand, describes the proportion of sequenced bases relative to the entire genome size. Standard genome sequencing protocols typically advocate for an average coverage depth of 30×. This recommendation stems from the observation that at this depth, the proportion of >4× coverage surpasses 99.21%, indicative of approaching saturation. Moreover, at this juncture, the number of heterozygous single nucleotide polymorphisms (SNPs) tends to plateau. Research indicates that sequencing at an average depth of 15× achieves saturation with pure SNPs, while 30× coverage suffices for heterozygous SNPs.
Is Higher Sequencing Depth Always Preferable?
In high-throughput deep sequencing, sequencing depth directly impacts the accuracy of each base. When analyzing a homogenous specimen where each base exists in only one form, excessively high sequencing depth may not be essential for optimal results. Conversely, in heterogeneous specimens, maintaining the same sequencing depth may result in fewer genome read lengths covering the rarer cells.
Considering the impact of sequencing depth on the accuracy of genetic testing, one might argue for a higher sequencing depth requirement. Analogously, we can draw parallels to our daily lives: just as the size of our living space affects our comfort level, cost is also a crucial consideration. Therefore, we strive for an optimal balance within our budget. Similarly, within a certain range, increasing sequencing depth enhances assay accuracy. However, escalating sequencing depth indefinitely leads to diminishing returns: while costs soar, improvements in accuracy become marginal.
How to Determine Sequencing Depth
Choosing the appropriate sequencing depth hinges on various factors. In cancer research, for instance, the required sequencing depth escalates for tumors with low purity, high polyclonality, and applications necessitating heightened sensitivity, such as identifying low-frequency clones. Typically, sequencing depths in cancer research span from 80x to several thousand times. Hence, from a cost-effectiveness standpoint, routine genetic testing for patient-targeted therapy guidance prioritizes optimal depth rather than maximal depth, reserving very high sequencing depths for specific scenarios.
For NGS testing of tumor tissue samples, the effective sequencing depth should exceed 500x, while for plasma-free DNA specimens, it should surpass 1000x.
Sequencing depth and coverage partly determine the confidence level associated with identified variants at specific base positions. Higher depth and coverage entail more reads covering each base, thereby bolstering confidence in the base calls.
Recommended depths vary based on the sequencing method:
- Whole genome sequencing (WGS): Typically 30× to 50×, contingent upon the application and statistical model.
- Whole exome sequencing: The suggested depth is around 100×.
- RNA sequencing: Detecting rare expressed genes necessitates higher coverage and depth.
- ChIP-Seq: Advised depth is around 100×.
Deep Sequencing Technology for Oncology
Tumor specimens in oncology research present a complex landscape, often comprising a blend of normal cells alongside multiple subclones of tumor cells. Addressing this intricacy requires a nuanced understanding of sequencing depth and its pivotal role in enhancing the accuracy and sensitivity of assays.
Considerations for Enhanced Sequencing Depth:
- Tumor Purity: The presence of normal cells within tumor specimens poses a challenge for accurate mutation detection. Tumors with 50% normal cells necessitate a doubling of sequencing depth to achieve the same confidence level as samples devoid of normal tissue.
- Tumor Heterogeneity: Polyclonality is a hallmark of advanced tumors, underscoring the need for deeper sequencing to capture the diversity of clone types present. The depth of sequencing must align with the complexity of the tumor's clonal architecture to ensure comprehensive variant detection.
- Required Sensitivity: Unveiling rare clones, particularly those constituting a mere 1% of the original tumor, is critical for understanding mechanisms of drug resistance and disease progression. Achieving this level of sensitivity mandates sequencing depths significantly deeper than conventional standards, often exceeding 100x coverage.
By meticulously addressing these factors, deep sequencing technology elevates the sensitivity and precision of oncological assays, empowering researchers to glean invaluable insights into tumor biology and therapeutic resistance mechanisms.
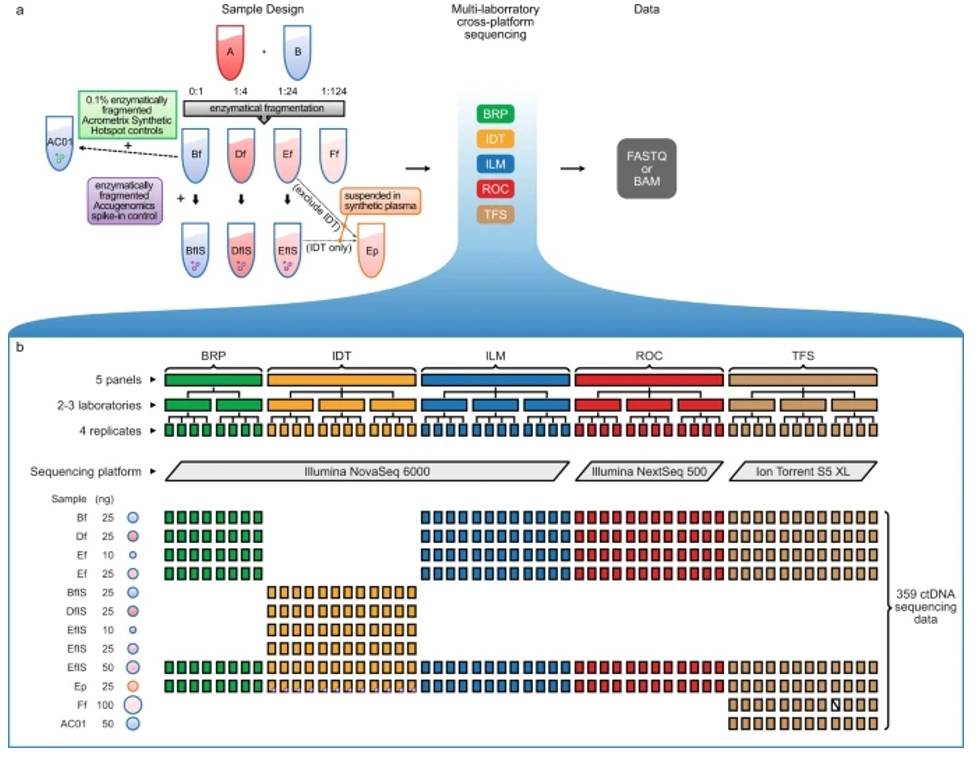 Ultra-deep sequencing data from a liquid biopsy proficiency study demonstrating analytic validity. (Gong et al., 2022)
Ultra-deep sequencing data from a liquid biopsy proficiency study demonstrating analytic validity. (Gong et al., 2022)
Deep Sequencing Technology in Virology
- Influenza Virus
To investigate potential pathogens and their interactions in patients afflicted with influenza A viruses, Kuroda M et al. conducted a comprehensive analysis. They extracted total RNA from the lungs of patients succumbed to viral pneumonia caused by H1N1, synthesized cDNA, and performed de novo sequencing. The study revealed that over 98% of the sequences belonged to the human genome, while H1N1 virus and bacterial sequences constituted 0.850% and 0.005%, respectively.
Moreover, the study unveiled two amino acid quasi-species phenomena in specific sites of the H1N1 virus's hemagglutinin antigens, Sa and Ca, corroborated by fluorescence quantitative PCR. Additionally, the identification of an unprecedented bacterial sensing agent and histologically unobserved bacterial infection underscored the significance of the findings.
These discoveries shed light on previously unrecognized bacterial infections, suggesting a broader contribution of potential pathogens, including Streptococcus pneumoniae, to the exacerbated morbidity and mortality associated with influenza A viral infections. Consequently, the widespread adoption of deep sequencing as a rapid and cost-effective pathogen detection tool ensued.
- Application in Unknown Virus Detection
Rotavirus stands as a leading cause of acute gastroenteritis in infants worldwide, with vaccination representing a pivotal preventive measure. Quality control of vaccines necessitates the detection of exogenous factors, crucial indicators of vaccine safety. In 2010, deep sequencing technology facilitated the detection of porcine circovirus.
This detection marked a significant event in vaccine safety regulation, akin to the intussusception incident, underscoring the indispensable role of deep sequencing in safeguarding public health.
- Application in Plant Viruses
Next-generation sequencing (NGS) has emerged as a cornerstone in identifying unknown pathogens in various plant species. For instance, Li et al. utilized NGS to analyze small RNAs from tomato samples exhibiting diverse symptoms. By enriching and assembling virus-derived small interfering RNAs (vsiRNAs), they identified a complete genome sequence of Potato spindle tuber viroid (PSTVd) and confirmed the presence of two Pepino mosaic virus (PepMV) strains in the samples from the United States. Furthermore, novel PepMV strains were discovered in Mexican samples, highlighting the utility of deep sequencing in uncovering novel plant viruses and strains.
Reference:
- Gong, B., Deveson, I.W., Mercer, T. et al. Ultra-deep sequencing data from a liquid biopsy proficiency study demonstrating analytic validity. Sci Data 9, 170 (2022).


 Sample Submission Guidelines
Sample Submission Guidelines
