How Intronic Variation Regulates Fruit Color Revealed through Pan-Genomic Mapping
Intronic Variation in Plant Genomics Research
A significant portion of the inherent diversity within plant populations arises within non-genetic coding regions, with a particular emphasis on intronic and intergenic regions. Understanding the regulatory mechanisms governing the variation within intronic regions and unraveling its impact on phenotypic traits stand as crucial challenges in the field of genetics research. Recent strides in genetic variation regulation, particularly in plants and animals, including human diseases, have shed light on the intricate ways in which intronic variation influences mRNA shearing and transcription.
Despite these advancements, our knowledge about the extent to which intronic variants can shape their non-coding roles and their specific effects on agronomic traits remains limited. Further exploration in this realm is imperative to comprehensively grasp the functional implications of intronic variation and unlock its potential contributions to understanding and manipulating plant characteristics for agricultural purposes.
In a recent study, the pan-genome of apple materials was meticulously mapped. This exploration not only unveiled the impact of copy number variations (CNVs) on gene expression but also brought to light crucial structural variations (SVs) that play a pivotal role in regulating fruit traits. This discovery holds significant importance in enhancing our comprehension of the molecular genetic mechanisms governing fruit traits, particularly those related to coloration. Additionally, the study has furnished valuable genetic resources that can prove instrumental in advancing apple breeding efforts.
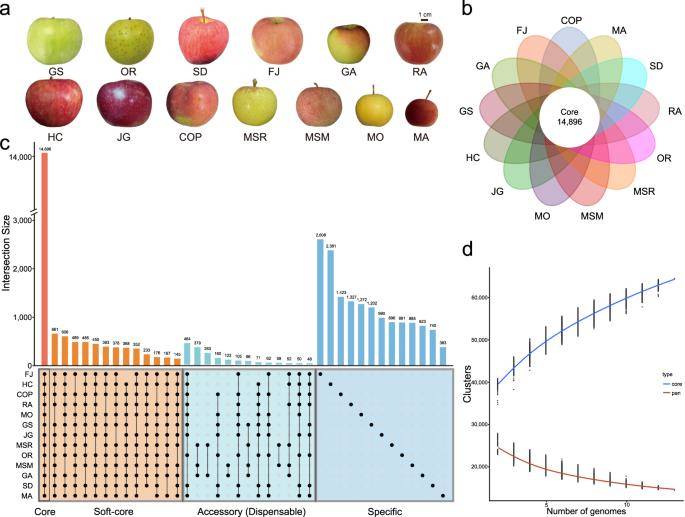 Pan- and core genome analysis of 13 apple accessions. (Wang et al., 2023)
Pan- and core genome analysis of 13 apple accessions. (Wang et al., 2023)
Varietal Traits and the Intricacies of Adaptive Evolution
This study utilized two wild apple species and eight cultivated varieties, chosen for their representative fruit traits. Employing PacBio SMRT sequencing and high-throughput chromosome conformation capture (Hi-C) assembly methods, genomes ranging from 661.83 to 668.75 Mb were obtained, with contig N50s between 24.06 and 39.17 Mb. Constructing 13 apple pan-genomes, including three published genomes, revealed core gene families comprising 32.10% to 48.72%.
To comprehensively assess genetic variation, 20,220 CNVs and 317,393 SVs were identified. Some CNVs showed increased gene expression with copy number elevation. Structural variant genome-wide association analysis (SV-GWAS) identified SVs linked to apple coloring. Notably, an LTR/Gypsy TE insertion in the kinase MMK2 intron 4 correlated with uncolored peel, while its absence marked colored peel germplasms. Further investigation revealed that this intronic variation influenced MMK2 expression by regulating its transcription into a non-coding RNA, impacting pericarp coloration. This sheds light on the nuanced role of intronic variation in kinase-like genes during apple domestication and breeding, advancing our understanding of adaptive evolution mechanisms in apples.
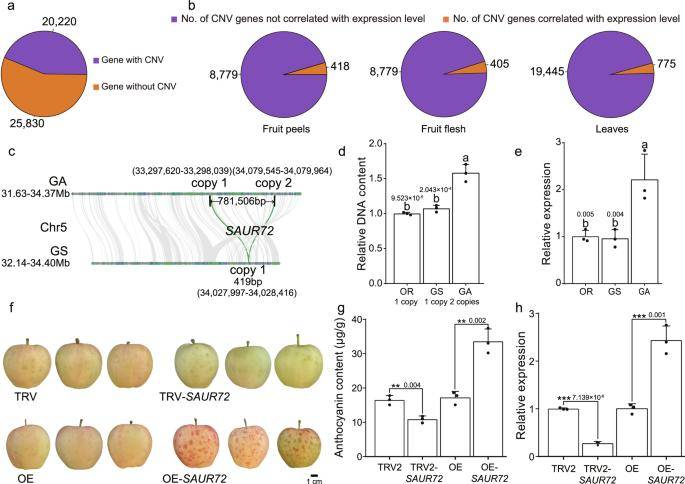 Gene CNVs are widespread and are associated with agronomic trait variation. (Wang et al., 2023)
Gene CNVs are widespread and are associated with agronomic trait variation. (Wang et al., 2023)
This study not only enhances our understanding of the evolution of the apple genome and key traits but also establishes vital platform resources and information for identifying crucial genes governing apple fruit traits and supporting genetic breeding efforts. The proposed intronic variation regulation mechanism and its associated findings also serve as valuable references for exploring genetic variation in other crops.
Reference:
- Wang, T., Duan, S., Xu, C. et al. Pan-genome analysis of 13 Malus accessions reveals structural and sequence variations associated with fruit traits. Nat Commun 14, 7377 (2023).


 Sample Submission Guidelines
Sample Submission Guidelines
