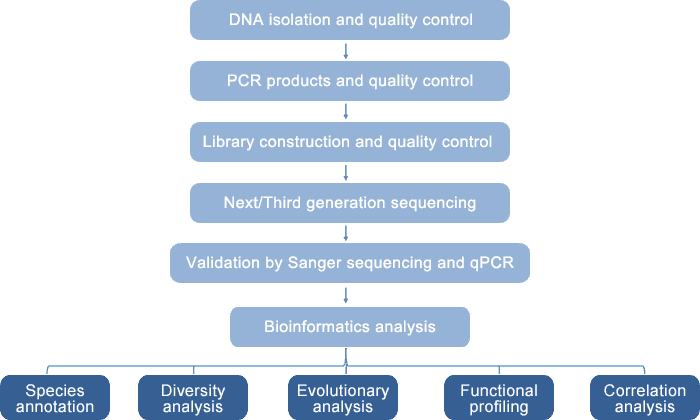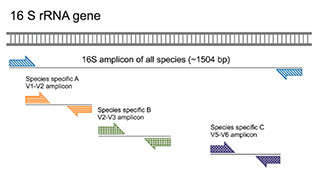
Our integrated prokaryotic diversity analysis platform can be used for profiling of prokaryotic community in your samples utilizing diverse molecular biology techniques. The principle of our prokaryotic diversity detection platform is based on the role of 16S rRNA as the molecular marker in prokaryotes. 16S rRNA gene has nine conserved regions and nine hypervariable regions. Among them, V4-V5 region is the best choice for bacterial diversity analysis and annotation due to its high specificity and large amount of database information. The two sets of primers (519F/915R and 344F/915R) offer the highest resolution for archaebacterial diversity analysis. This platform enables both short-read 16S rRNA sequencing by Illumina HiSeq/Roche 454 instruments and full-length 16S rRNA sequencing by PacBio SMRT sequencing or Nanopore sequencing. The generated sequencing data are validated by Sanger sequencing and q-PCR with genus- and species-specific primers. qPCR can also be used to quantify microbial species.
Based on this platform, we provide comprehensive prokaryotic diversity analyses for bacteria, archaea, actinomycetes. Bacteria and archaea are found in a broad range of habitats, such as soils, water, marshlands, and human body. Archaea even survive in harsh environments. The insight into prokaryotic community structure is an efficient way to understand the role of prokaryotes played in environments or human body, hence making use of prokaryotes or their products. Therefore, prokaryotic diversity analysis has been applied in a variety of fields, including basic research (such as prokaryotic evolution and epidemiologic studies), agricultural practice, environmental protection, as well as pharmaceutical, food, healthcare, and other industries. Additionally, we also have eukaryotic diversity analysis platform for eukaryotic samples (such as fungi, algae, protozoa).