Genome Annotation and Gene Prediction Service
Introduction
In the era of genomics, unraveling the mysteries of genetic information holds the key to groundbreaking discoveries in fields like medicine, agriculture, and biodiversity. One crucial step in this endeavor is genome annotation, which involves deciphering the functional elements within a genome. CD Genomics, a leading name in the field, offers an advanced Genome Annotation and Gene Prediction Service that aids researchers in extracting valuable insights from genomic data.
The Process of Genome Annotation
Genome annotation is a complex and multi-step process that involves the identification and annotation of various genomic features, such as genes, regulatory elements, and non-coding RNAs. CD Genomics, a leading genomics service provider, employs a systematic approach to ensure accurate and comprehensive genome annotation. In this section, we will delve into the process of genome annotation, highlighting the key steps involved.
Step 1: Data Acquisition and Preprocessing
The genome annotation process begins with the acquisition of raw genomic data, which can be obtained through various sequencing technologies such as next-generation sequencing (NGS) or third-generation sequencing. CD Genomics possesses state-of-the-art sequencing platforms and expertise to generate high-quality genomic data.
Once the raw sequencing data is obtained, it undergoes preprocessing steps to remove artifacts, adapter sequences, and low-quality reads. Quality control measures are implemented to ensure the reliability of the data, thus laying a solid foundation for subsequent analysis and annotation.
Step 2: Genome Assembly
The next step in the annotation process is genome assembly, where the raw sequencing data is reconstructed into a contiguous representation of the genome. CD Genomics employs advanced computational algorithms and assembly tools to accurately assemble the genome, taking into account factors such as repetitive regions, sequencing errors, and heterozygosity.
The assembled genome serves as the scaffold for subsequent annotation steps, providing the framework for identifying and characterizing genomic features.
Step 3: Gene Prediction and Structural Annotation
Gene prediction is a crucial step in genome annotation, as it involves identifying the locations and structures of protein-coding genes within the genome. CD Genomics utilizes a combination of computational algorithms and machine learning approaches to accurately predict genes.
These algorithms analyze various genomic features, including open reading frames (ORFs), codon usage patterns, and sequence conservation, to identify potential coding regions. The predicted genes are then annotated with information such as gene boundaries, exon-intron structures, and coding sequences.
Step 4: Functional Annotation
Functional annotation aims to assign biological function and significance to the annotated genomic features. CD Genomics employs a diverse range of approaches to enhance functional annotation, including comparative genomics analysis, homology searches against known protein databases, and functional enrichment analysis.
Comparative genomics analysis involves comparing the annotated genome with existing genomic databases to identify conserved regions and orthologous genes across species. This analysis provides valuable insights into the evolutionary relationships and functional conservation of genes.
Homology searches against known protein databases, such as RefSeq and UniProt, help in assigning putative functions to the predicted genes based on sequence similarity to characterized proteins.
Functional enrichment analysis involves identifying overrepresented gene ontology terms, pathways, and functional modules within the annotated gene set. This analysis provides a comprehensive understanding of the biological processes and pathways associated with the genes, shedding light on their functional roles.
Step 5: Manual Curation and Validation
To ensure the accuracy and reliability of the genome annotation, CD Genomics employs a rigorous manual curation process. Experienced biologists and bioinformaticians meticulously review and refine the automated predictions, incorporating their domain knowledge and the latest scientific literature.
Through this manual curation process, potential errors and inconsistencies in the automated annotations are rectified, and additional information, such as alternative splicing variants and non-coding RNA transcripts, are incorporated.
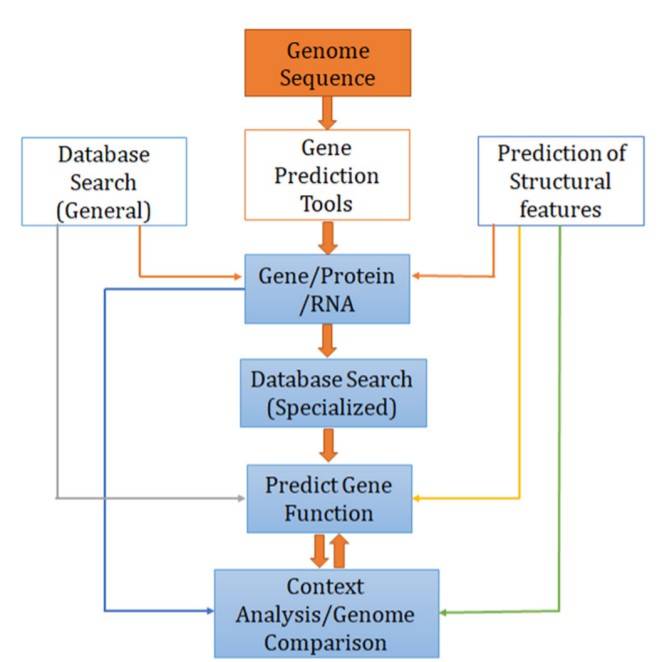
Genome Annotation Service Content
CD Genomics offers a comprehensive Genome Annotation and Gene Prediction Service that encompasses a wide range of analyses and deliverables. By utilizing state-of-the-art bioinformatics tools and leveraging their extensive knowledge in genomics, CD Genomics delivers accurate and actionable results to researchers.
Gene Prediction and Characterization
CD Genomics excels in gene prediction by employing advanced computational algorithms. Their team of experts identifies potential protein-coding genes and characterizes their structures, including coding sequences, untranslated regions, and alternative splicing variants. This comprehensive gene catalog serves as a foundation for functional studies and provides insights into the genetic basis of biological processes.
Non-Coding RNA Identification
Non-coding RNAs play crucial roles in gene regulation and other biological processes. CD Genomics employs specialized algorithms to identify various classes of non-coding RNAs, such as transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), and microRNAs (miRNAs). This comprehensive non-coding RNA annotation facilitates the understanding of regulatory networks and aids in the discovery of novel regulatory elements.
Functional and Structural Annotation
CD Genomics integrates functional and structural annotation approaches to unravel the complexities of the genome. By combining computational predictions with experimental evidence, they identify key regulatory elements, such as promoters, enhancers, and transcription factor binding sites. This information sheds light on the gene regulatory networks and provides researchers with a deeper understanding of the genome's functional architecture.
Manual Curation and Expert Review
CD Genomics emphasizes the importance of manual curation and expert review in ensuring the accuracy and reliability of genome annotation. Their team of experienced biologists and bioinformaticians meticulously scrutinizes the automated predictions, verifying and refining the annotations based on their domain knowledge and the latest scientific literature. This manual curation process ensures that the genome annotations provided by CD Genomics are of the highest quality and can be confidently utilized for downstream analyses and interpretation.
Comparative Genomics Analysis
CD Genomics employs comparative genomics analysis to enhance the functional annotation of genomes. By comparing the newly annotated genome with existing databases, they identify orthologous genes and conserved genomic regions across species. This comparative approach enables researchers to gain insights into the evolutionary relationships, functional conservation, and divergence of genes, contributing to a deeper understanding of the genome's biological significance.
Functional Enrichment Analysis
To uncover the functional implications of the annotated genes, CD Genomics conducts functional enrichment analysis. This analysis involves the identification of overrepresented gene ontology terms, pathways, and functional modules within the annotated gene set. By elucidating the biological processes and pathways associated with the genes, CD Genomics enables researchers to unravel the underlying mechanisms governing complex biological phenomena.
Genome Annotation Deliverables
CD Genomics ensures that researchers receive comprehensive deliverables that empower them to explore and interpret genome annotations effectively. These deliverables include:
Genome Annotation Report
CD Genomics provides a detailed and user-friendly report summarizing the genome annotation results. This report includes information on gene predictions, non-coding RNAs, functional annotations, regulatory elements, and comparative genomics analysis. The report is tailored to meet the specific requirements of the researchers, providing them with a comprehensive overview of the annotated genome.
Genome Browser Tracks
CD Genomics generates genome browser tracks that visualize the annotated features and their associated experimental evidence. Researchers can easily navigate and explore the genome annotations using popular genome browsers, enabling them to examine gene structures, regulatory elements, and other functional elements in the context of the genome sequence.
Data Files and Databases
CD Genomics provides researchers with the raw data files, intermediate results, and fully annotated genome databases. These resources enable researchers to perform their own in-depth analyses, integrate the annotations with other datasets, and extract valuable insights for their specific research questions.
Advatages of our Genome Annotation and Gene Prediction Service
Accurate and comprehensive annotation through advanced computational algorithms and manual curation.
High-quality data analysis using state-of-the-art sequencing technologies and rigorous quality control.
Customized annotation strategies tailored to specific research needs.
Integration of multi-omics data for a holistic understanding of gene expression and regulation.
Rapid turnaround time without compromising quality.
Enhanced data visualization and interpretation through intuitive tools.
Post-annotation support for guidance and assistance in downstream analysis.


 Sample Submission Guidelines
Sample Submission Guidelines
