The Origin and Evolution of Endogenous Retroviruses
Retroviral particles house the complete viral genome in the form of RNA. Upon entering a target cell, this RNA can undergo a transformation known as reverse transcription, converting it into double-stranded DNA, which is subsequently integrated into the host genome. This integrated viral DNA sequence is termed a provirus, encompassing essential elements like a promoter, regulatory segments, and coding sequences for both structural proteins and enzymes.
Typically, retroviruses infect somatic cells, but there exists a probability that a retrovirus may infect germline cells, thereby entering the host's gene pool. At this juncture, the provirus evolves into an endogenous retrovirus (ERVs). The destiny of ERVs within a population hinges on the forces of genetic drift and natural selection. In essence, ERVs represent genetic loci originating from exogenous retroviruses that have assimilated into the host genome through the process of endogenization. These entities retain the title of ERVs regardless of their capacity to continue expressing infectious viral particles. Over millions of years, many ERV sequences have lost their ability to produce viral particles due to accumulated mutations.
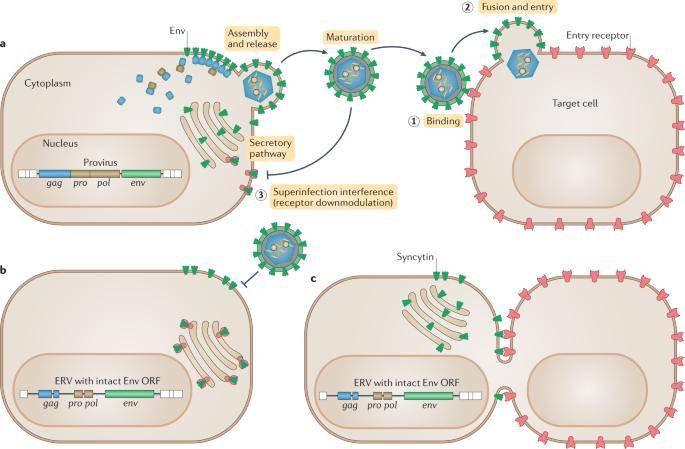 Env exaptation and the relationship between ancient viral functions and current genome functions. (Johnson Welkin E, 2019)
Env exaptation and the relationship between ancient viral functions and current genome functions. (Johnson Welkin E, 2019)
Diversity of Endogenous Retroviruses (ERVs)
Endogenous retroviruses share a common genome structure, facilitated by the processes of reverse transcription and integration. These processes result in the formation of proviral sequences ranging from 5,000 to 10,000 base pairs in length within the host genome, typically characterized by the presence of long terminal repeat sequences (LTRs) at both ends. These LTR sequences serve as essential elements, encompassing promoters for proviral expression, regulatory components, and cis-acting structural domains crucial for integration.
The genetic content of retroviruses typically comprises:
- gag: responsible for encoding the structural proteins that constitute the viral particle's core.
- pro: tasked with encoding the viral protease.
- pol: housing the genes for reverse transcriptase and integrase.
- env: responsible for encoding glycoproteins essential for facilitating fusion and cellular entry.
The LTR's terminal repeat sequence is typically divided into three segments, ordered from the 5' end to the 3' end: U3, R, U5.
At the 5' end, the junction between U3 and R marks the initiation of transcription, while at the 3' end, the junction between R and U5 denotes the termination of transcription.
The U3 segment is typically within the range of 190 to 1,200 base pairs and contains an array of structural elements, including promoters, enhancers, and other regulatory motifs. These elements have the capacity to interact with host cell regulatory factors, thereby exerting control over the protovirus's gene expression.
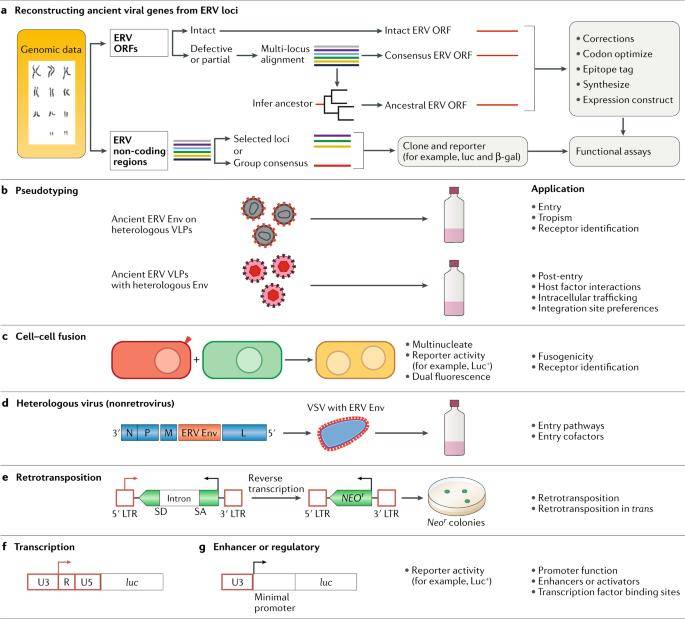 Reconstructing and analysing ancient endogenous retrovirus genes. (Johnson Welkin E, 2019)
Reconstructing and analysing ancient endogenous retrovirus genes. (Johnson Welkin E, 2019)
Ancient Retroviruses: Insights into Host Evolution
Endogenous retroviruses (ERVs) stand as genomic remnants of bygone retroviruses, offering a unique window into the co-evolution of retroviruses and their host genomes. They are invaluable tools for unraveling the impact of retroviruses on host evolution.
Take, for instance, HERV-K (HML-2), an ERV present in the human genome, the Neanderthal genome, and the Denisovan genome. Its ubiquitous presence suggests an ancestral retroviral origin within the human lineage. In contrast, it remains unfixed in the gorilla genome, residing in a genetically segregated state with a relatively complete sequence. This indicates ongoing activity in gorillas, with these retroviruses retaining the potential to generate new infectious viral particles.
ERVs affiliated with spumaviruses, discovered in marine animals, provide insights into a deeper historical context, suggesting that these viruses infected marine animals as far back as the Paleozoic era. The identification of as-yet-unfixed ERVs, like CrERV-gamma in deer, can shed light on the early stages of endogenization. Notably, in koalas, the retrovirus KoRV exhibits both endogenized, non-pathogenic sequences and sequences yet to undergo endogenization, which could potentially be pathogenic. Analyzing these cases aids in comprehending the endogenization process and the virus's influence on host genome evolution.
ERVs can also serve as molecular clocks, enabling the estimation of integration timing and providing insights into the emergence and dissemination of ancient retroviruses.
Highly Conserved env Genes: Insights into Evolution
Among the most ancient complete env genes discovered, we find those within the percomorf gene of fin-spined fish and the primate HEMO gene, which have persisted for hundreds of millions of years. Their enduring presence suggests remarkable conservation and enduring negative natural selection, hinting at their potential vital cellular functions.
For instance, the percomorf gene's conservation may not stem from its ability to resist viral invasions but from its capacity to induce receptor-mediated membrane fusion. On the other hand, while not associated with membrane fusion, the HEMO gene expresses the full-length Env protein, subject to cleavage by cytosolic proteases. A portion of the cleaved product is secreted outside the cell, detectable in the blood of pregnant women and in the blood and tissues of the placenta. Despite these intriguing findings, the precise physiological role of this phenomenon remains enigmatic.
Env proteins encoded by most retroviruses can be broadly categorized into two types: gamma-type and beta-type.
- Gamma-type predominates in vertebrates.
- Beta-type is more prevalent in mammals.
The current emphasis on the domestication of Env genes has primarily focused on the gamma type. This choice likely derives from the fact that gamma-type env genes are less prone to inducing cell fusion, simplifying the process of domestication by the host.
Genomic Imprinting of Domestication
Within ERV sequences, the env gene, responsible for conferring host resistance against viruses, is typically accompanied by intermittent gag, pro, and pol genes. This dual observation implies two crucial aspects: the env gene is highly conserved and subject to sustained negative natural selection, while several other genes have accumulated a significant number of random substitutions.
Various methods for assessing natural selection can be employed, such as the dN:dS ratio, which measures the ratio of non-synonymous to synonymous substitutions. A ratio less than 1 signifies negative natural selection, equality to 1 denotes random genetic drift, and greater than 1 indicates positive natural selection. Negative and positive natural selection can coexist within a single gene; certain codons may be under positive selection, while others may experience negative selection to maintain essential protein structure and function. Notably, for both ERVs, percomorf and HEMO, the dN:dS ratio is less than 1, suggesting they are indeed subject to negative natural selection. In contrast, fv1 has been under sustained positive natural selection, along with target-specific intrabranchial selection pressure.
If the Env gene plays a vital role in species development, it should remain highly conserved and experience enduring negative selection pressure. However, if its primary function is to inhibit exogenous retroviral invasion, the selective pressure against antiviruses should exhibit cyclical patterns, intensifying with the emergence of exogenous viruses and waning or altering as exogenous viruses go extinct. The study's results confirm this hypothesis, with syncytins, linked to placental formation development, originating as early as 12 to 80 million years ago, while certain Env genes focused on resisting viral invasion are relatively recent, with emergence events occurring later than 20 million years ago.
Domestication of Non-Coding Elements
Proto-viruses and ERVs have the capacity to influence the regulated expression of genes located within a few kilobases of their surroundings. ERV LTRs can serve as promoters or sites for transcription factor binding, potentially contributing long non-coding RNAs (lncRNAs) that act as regulatory sequences. This dynamic acquisition and loss of ERV regulatory sequences during evolution enable even closely related species to manifest significant phenotypic diversity.
Several key features of ERVs can contribute to the evolution of host gene regulatory networks.
- LTR sequences harbor a high density of regulatory elements, encompassing promoters and transcription factor binding sites. The nature of these sequences reflects the properties of the original viruses at the time of integration, including factors such as host and tissue selectivity.
- Retroviral integration, while not entirely target sequence-specific, tends to favor transcription units and regions containing promoters in the host genome. This non-random distribution of insertion sequences generated during endogenization is subject to various evolutionary forces. Some sequences are subjected to genetic drift or negative selection, gradually dwindling in the population, while others experience natural selection and become fixed in the population, persisting over time.
- Similar LTR sequences can induce chromosomal ectopic recombination or gene conversion.
- Solo-LTR retains only the retroviral LTR sequences, shedding coding sequences and circumventing epigenetic modifications of the genome caused by these sequences. This allows for the preservation of the regulatory potential of LTR sequences.
Reference:
- Johnson, Welkin E. "Origins and evolutionary consequences of ancient endogenous retroviruses." Nature Reviews Microbiology 17.6 (2019): 355-370.


 Sample Submission Guidelines
Sample Submission Guidelines
