Mapping of Oncogenic Structural Variants in Gastric Cancer Uncovers 27 Potential Driver Sites
Gastric cancer (GC) is a prevalent form of cancer worldwide, ranking as the fifth most common. Understanding the genetic and epigenetic changes associated with gastric cancer drivers is crucial for targeted therapies. Key driver events like ERBB2 and VEGFA alterations have been identified as important therapeutic targets. Extensive genomic analyses of gastric cancer have unveiled multiple driver events and mutational processes, including mutational signatures that relate to geographic, epidemiologic, and histologic factors.
Although the role of structural variants (SVs) in gastric cancer is acknowledged, their specific patterns and mechanisms are not yet fully understood. Certain significant driver SVs, such as the CLDN18-ARHGAP fusion gene and KLF5 enhancer amplification, have been previously reported. Recent studies have examined the patterns of structural variation (SV signatures or rearrangement signatures) across various human cancer types. However, these studies have not adequately explored the clinical and epidemiologic contexts that underlie these patterns.
Oncogenic Structural Variants Map
A study titled " Oncogenic structural aberration landscape in gastric cancer genomes" was published in Nature Communications. In this study, the researchers conducted a comprehensive analysis of 170 gastric cancer cases to investigate the landscape of oncogenic structural aberrations in gastric cancer genomes. By examining the entire genomes of these cases, the team identified six distinct rearrangement signatures (RS) and pinpointed 27 potential driver SV sites that contribute to the development of gastric cancer. The findings shed light on the intricate genetic mechanisms underlying gastric cancer and offer valuable insights for future therapeutic approaches.
In this study involving 170 gastric cancer genomes, researchers discovered a total of 49,059 structural variations (SVs). These SVs comprised 22,179 deletions, 11,234 tandem repeats, 8,534 inversions, and 7,112 translocations. Smaller SVs, around 100 kilobases in size, involving deletions and tandem duplications, exhibited higher SVAF (SV allele frequency), indicating their early acquisition. Tandem repeats with substantial copy number variations demonstrated significantly higher SVAF, displaying a size-dependent trend, which suggests their potential as cancer drivers. On the other hand, translocations exhibited lower average SVAF compared to other types of SVs, confirming their acquisition through genome-wide hypomethylation during cancer progression.
By utilizing SVAF, the researchers aimed to identify driver SVs on a genome-wide scale in gastric cancer. They found that SV hotspots frequently coincided with high SVAF genes associated with gastric cancer oncogenes and fusion genes. Additionally, hotspots of high SVAF tandem repeats and inversions were observed at known oncogene loci like ERBB2 and CCNE1, as well as other loci.
The study specifically focused on these two types of SVs and identified 27 genomic segments as candidate driver SV hotspots. These segments met the following four criteria: 1) detected in more than 5% of cases; 2) not located within common fragile sites (CFS) or active transposons; 3) contained five or more cases with high SVAF (>0.4); and 4) the number of SV cases had significantly higher copy numbers compared to cases without SVs, indicating increased ratios.
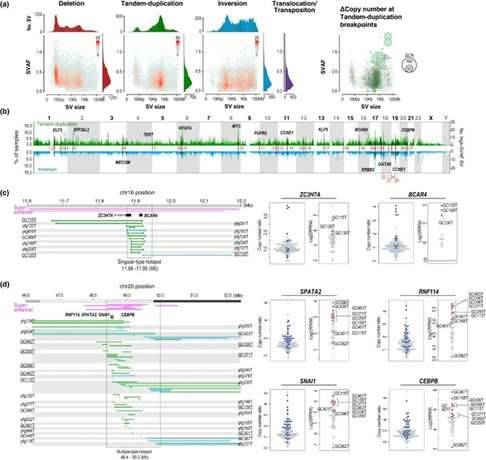 Structural variants in 170 gastric cancer (GC) genomes. (Saito-Adachi et al., 2023)
Structural variants in 170 gastric cancer (GC) genomes. (Saito-Adachi et al., 2023)
To investigate the molecular and epidemiological aspects of SV accumulation, researchers conducted an analysis wherein they extracted SV features that represented the tendencies of SVs. The study identified six distinct SVs, referred to as RSs, based on 80 Signal SV classification features and additional categories. Each RS displayed a characteristic chromaticity distribution.
By utilizing unsupervised hierarchical clustering, the researchers classified a total of 170 cases into seven RS subtypes: RS1-6 and RS2/6. Six subtypes consisted of a single RS as the primary feature, while the last subtype contained both RS2 and RS6. RS1 and RS4 were predominantly composed of deletion-type SVs. RS2 comprised sparsely distributed small SVs, RS6 consisted of densely distributed large SVs, and RS3 exhibited large (ranging from 1kb to 1mb) tandem overlaps in regions characterized by active chromatin. Cases with RS6 and RS2/6 subtypes displayed a poorer prognosis.
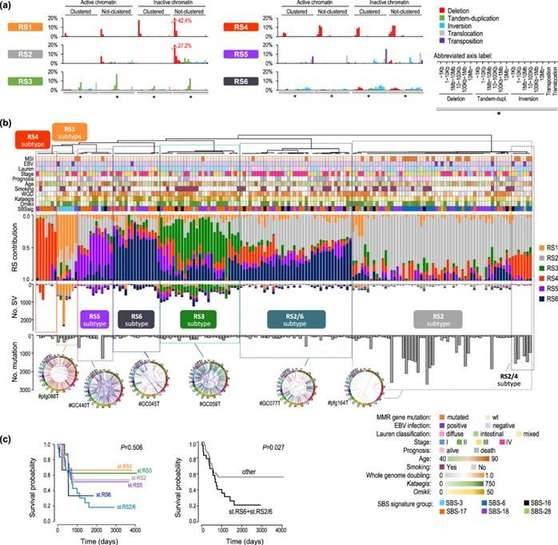 GC Structural rearrangement signatures (RSs). (Saito-Adachi et al., 2023)
GC Structural rearrangement signatures (RSs). (Saito-Adachi et al., 2023)
SV clusters, known as SVCs, are frequently observed in cancer genomes and are linked to the amplification of oncogenes. In gastric cancer, approximately 36.6% of SVs form 3,457 SV clusters, with variations in their distribution across different RS subtypes. Within the RS2 subtype, less than 20% of SVs belong to SV clusters, whereas the RS3 and RS6 subtypes exhibit more than 50% of SV clusters. To understand these clusters better, researchers categorized all 3,457 SV clusters based on various factors such as SV type, size, distribution, and SVAF. They also annotated the proposed molecular mechanisms.
The findings revealed that CFS-like SV clusters were characterized by recurring deletions of varying sizes within the same region, accompanied by a gradual decline in copy number. Out of the 793 CFS-like SV clusters, 38.9% overlapped with validated CFS segments documented in HumCFS. Notably, the RS4 isoforms primarily consisted of CFS-like SV clusters (83.3%). On the other hand, L1-retrotransposon-type SV clusters were typically located within 1.0 kb of the LINE1 element and were frequently observed in RS5 isoforms (24.9%).
Additionally, the analysis unveiled other complex SV clusters that could not be classified. These clusters exhibited breakpoints with high SVAF at the edges of the amplicon, while a set of lower SVAFs clustered within the amplicon. In some instances, these amplicons were interconnected at distances exceeding 2 Mb. The researchers referred to the former as self-joining amplicons and the latter as assembled amplicons. The number of these complex SV clusters correlated with the count of "aggregated large-scale SVs" within the RS6 signature.
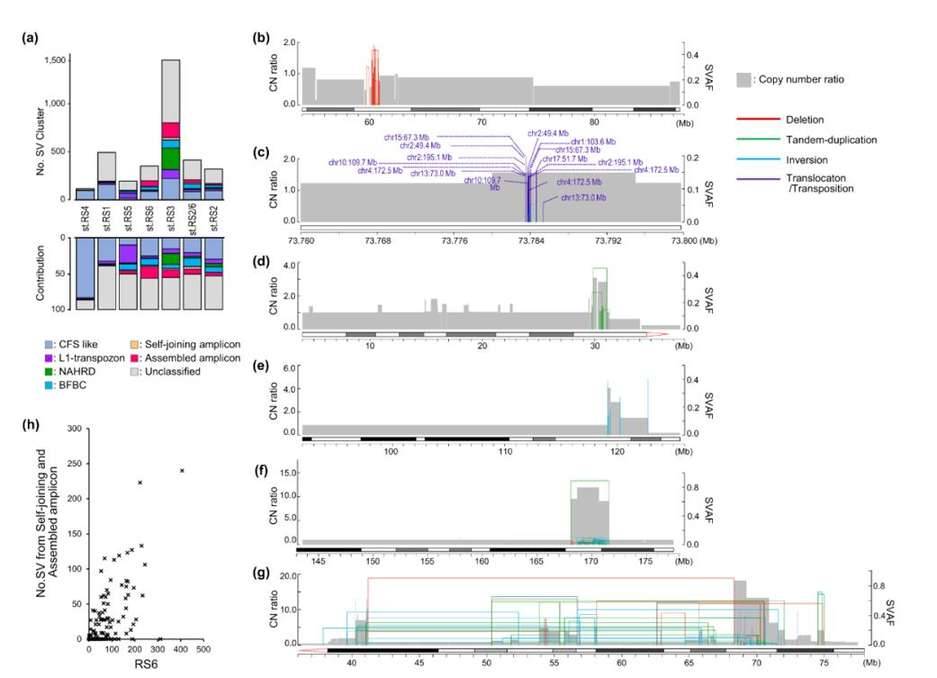 Molecular classification of SV clusters (SVCs). (Saito-Adachi et al., 2023)
Molecular classification of SV clusters (SVCs). (Saito-Adachi et al., 2023)
Summary
The researchers examined the entire genome of 170 individuals with gastric cancer and successfully identified recurring structural variations (SVs) that could potentially drive the development of gastric cancer. Their focus was on tandem duplications and inversions that led to segmental amplification. From this analysis, they pinpointed 27 hotspot locations, which they further classified into two types: monotypic and multiplex.
The monotypic type was characterized by gene fusions, disruption of oncogenes, and the duplication of gene regulatory elements at a low level. On the other hand, the multiplex type emerged as a result of a combination of different SV types, leading to a higher level of copy number amplification. In these instances, the number of SVs varied significantly and occasionally formed complex SV clusters. These clusters were characterized by the accumulation of subclonal SVs, which gradually amplified through inaccurate repair of double-stranded DNA breaks and the segregation of chromosomes. These distinct characteristics indicate a specific distribution pattern of driver SVs.
Reference:
- Saito-Adachi, Mihoko, et al. "Oncogenic structural aberration landscape in gastric cancer genomes." Nature Communications 14.1 (2023): 3688.


 Sample Submission Guidelines
Sample Submission Guidelines
