The 16S rRNA gene, a DNA sequence encoding rRNA present in the genome of all bacteria, is a pivotal molecular marker in microbial ecology due to its universal existence and unique evolutionary characteristics.
Given that the 16S rRNA gene remains conserved within bacteria and harbors hypervariable regions that can offer species-specific sequence signatures, it becomes an extensively leveraged tool for bacterial identification and phylogenetic studies. Its attributes of rapid processing, cost-effectiveness, and high precision make it particularly appealing. The use of the 16S rRNA sequencing technique has broadly permeated basic research and fields including medical microbiology, forensic science, agricultural and industrial microbiology.
Service you may intersted in
Fundamental Research
16S rRNA sequencing plays a vital role in general investigations, including taxonomy, ecological studies, and evolutionary research.
Primarily, 16S rRNA sequencing enables the identification of unknown microorganisms' species and subspecies, facilitating microbial taxonomy. The homology of 16S rRNA gene sequences has provided one of the most rapid methods for determining species' taxonomic status. Presently, GenBank and EzBiocloud databases hold a vast amount of 16S rRNA gene sequences originating from type strains of various prokaryotes. These sequences can swiftly and accurately classify unknown bacteria through online BLAST alignments. Additionally, 16S rRNA sequencing is widely used in characterising microorganisms from multiple environments such as rivers, oceans, and soil.
Moreover, 16S rRNA sequencing finds extensive use in discerning and comparing phylogenetic relationships among different species and strains, assisting in the construction of a phylogenetic tree of life. It can inform the evolutionary distance and common ancestors among species, investigate the process of species formation and differentiation, and explore genetic divergence and population structure between species.
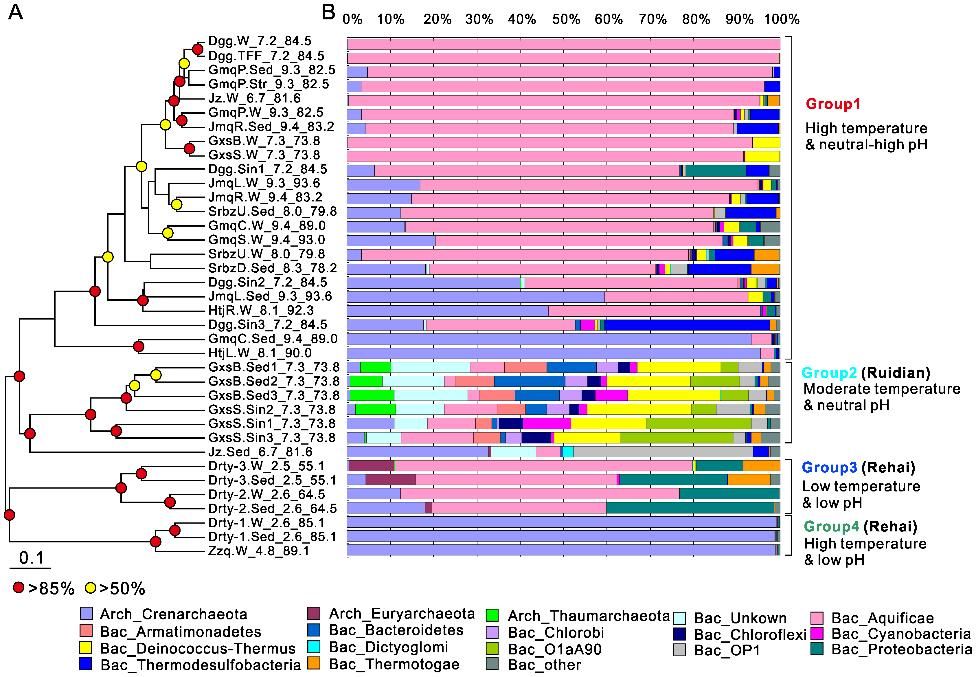 Figure 1. Microbial community composition grouped by pH and temperature. (Hou et al., 2013)
Figure 1. Microbial community composition grouped by pH and temperature. (Hou et al., 2013)
Furthermore, 16S rRNA sequencing provides high-throughput microbial taxonomic information, empowering ecologists to comprehend the diversity and composition of microorganisms in different environments. By sequencing the 16S rRNA from diverse environmental samples, such as soil, water bodies, air, and intrabiological samples, we can compare the structure and functions of microbial communities within diverse ecosystems and identify the roles and ecological niches of specific microbial communities. Besides, analysis of microbial community 16S rRNA sequences can elucidate key ecological processes within an ecosystem, including carbon and nitrogen cycles, thereby aiding scientists in further investigation.
Through the comparative examination of microbial communities in various environments, we can understand how microorganisms adapt to their surroundings and the symbiotic relationships among them. It helps investigate the evolutionary interplay between hosts and associated symbiotic microorganisms, such as the co-evolution of gut microbiota and hosts. By monitoring changes in the microbial communities within an ecosystem, we can assess the impact of environmental pollution, biological invasions, and other disruptive factors on the ecosystem's health, thereby assessing its stability and sustainability.
Medical Field
The application of 16S rRNA sequencing primarily spans disease diagnosis and monitoring, personalized healthcare, drug development, and the control and prevention of infectious diseases within the medical field.
16S rRNA sequencing is employed to detect bacteria and archaea in clinical samples, facilitating the diagnosis of bacterial infections, especially those caused by rare or challenging-to-culture pathogens. In clinical practice, by comparing the 16S rRNA sequences from patient samples with bacterial 16S rRNA sequences in a database, the causative agents of infections can be accurately identified. Qian and colleagues, by comparing the blood microbiome composition of 58 Parkinson's disease patients and 57 healthy individuals via 16S rRNA sequencing, have uncovered possible significant associations between the genera Paludibacter and Saccharofermentans and Parkinson's disease.
The use of 16S rRNA sequencing offers critical insights into patients' microbial communities, allowing for the prediction of drug metabolism and side effects. Furthermore, this sequencing technique can aid in the discovery of microbial markers associated with various diseases, providing potential targets for new drug development or evaluating drug efficacy. In a notable study, He and colleagues utilized both 16S rRNA sequencing and metabolomics analyses in tandem to identify biological markers associated with drug-induced liver injury. They constructed a co-linearity network diagram of key microbial metabolites. The results implied positive correlations between corticosterone, prostaglandin I1, Bioyclo prostaglandin E2, and oleanolic acid with the genera Blautia and Ralstonia. Interestingly, negative correlations were observed with the genus Veillonella. These findings expand our understanding of microbiome-drug interactions and provide potential avenues for therapeutic advancements and drug safety evaluation.
In the face of infectious diseases, the analysis of pathogenic microorganisms' 16S rRNA sequences could offer valuable insights into the routes of disease transmission and variation, thereby aiding in the timely implementation of strategies for infectious disease control. A study conducted by Morel and colleagues incorporated the evaluation of 32,948 clinical samples from 18,056 patients. By utilizing technologies such as 16S rRNA sequencing, they identified 1,192 instances of infection, providing a robust data foundation for future intervention measures.
Agriculture
The application of 16S rRNA sequencing in agriculture spans across several facets including soil health, crop growth, and disease control.
Assessments of soil health can be profoundly informed by monitoring alterations in soil microbial communities, based on data derived from 16S rRNA sequencing of these micro-ecosystems. This information serves as reference for soil management interventions. Moreover, the functional attributes of microorganisms residing in the soil can be elucidated via 16S rRNA sequences. This insight into the types and functionalities of beneficial microorganisms can optimize soil management practices, thereby enhancing soil fertility and crop yield.
The technique can also be employed to isolate microbial strains with positive traits. The application of these probiotics in soil or seed treatments can stimulate crop growth, fortify disease resistance, and augment yields. Conversely, pathogenic microbes can be identified, permitting predictions to be made and preventative measures to be enacted for crops, thus offering effective management strategies for disease control.
Furthermore, 16S rRNA sequencing can be used to monitor the response of soil microbial communities to environmental pollution, thereby providing scientific evidence for environmental protection and soil rehabilitation strategies.
Industrial
In the realm of industrial microbiology, 16S rRNA sequencing can serve as a pivotal tool for tasks such as microbial strain screening, fermentation process optimization, and quality control of products.
Sequencing the 16S rRNA allows for the discovery of novel microbial strains with specific metabolic traits or bioactivities, thus facilitating the development of new products or the enhancement of existing ones within industrial production. Moreover, predicting the metabolic pathways and biosynthesis capabilities of microbial strains can reveal potential for the production of novel bioactive substances.
During fermentation processes, utilizing 16S rRNA sequencing for monitoring the dynamic shifts in microbial communities can provide essential insights. Evaluating the competitive intricacies and operational mechanisms of various microbial strains during fermentation can assist in amplifying product yield and quality. Further benefits can be reaped through the identification and genetic manipulation of microbial strains with superior fermentation performance, thus bolstering both efficiency and yield.
In the context of biological treatment of industrial waste–encompassing wastewater, exhaust gases, and solid waste– 16S rRNA sequencing can enable the screening of microbial strains capable of wastage degradation, thereby facilitating resource recycling and environmental protection. Furthermore, the discovery of microbial strains with the potential for bioenergy production—namely the generation of biohydrogen, biomethane, and bioethanol—can spur industrial growth.
Forensic Science
In the realm of forensic science, 16S rRNA sequencing plays a pivotal role and is widely applied in areas such as identity verification, criminal investigations, and forensic pathology.
16S rRNA sequencing enables the identification and analysis of microbes found in human tissue samples (like blood, tissue, or bone), thus facilitating identity confirmation or validation. By implementing 16S rRNA sequencing on microbial samples collected from crime scenes, one can analyze the microbial community present in these specimens. Subsequently, comparison with microbial communities inherent to suspects' personal items, like mobile phones or clothing, can assist in pinpointing suspected individuals. In circumstances where sufficient human DNA samples are unattainable, sequencing the 16S rRNA of microbial communities can garner genetic information of victims or suspected criminals.
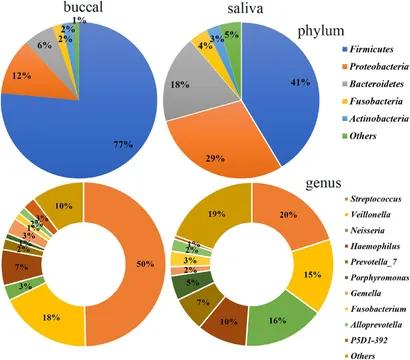 Figure 2. Bacterial community composition of the saliva and buccal mucosa at the phylum and genus levels. (Wang et al., 2022)
Figure 2. Bacterial community composition of the saliva and buccal mucosa at the phylum and genus levels. (Wang et al., 2022)
In forensic pathology, for unidentifiable corpses or remains, 16S rRNA sequencing helps in deciphering microbial DNA present within the body tissues, thus supplementing corpse identification and validation. The 16S rRNA sequencing of microbial communities in family members or identity-confirmed individuals can enable kinship evaluation, thereby contributing additional insights for forensic kinship identification. Further, the 16S rRNA sequencing of the corpse tissue samples can shed light on variations in microbial communities before and after death, thus aiding the determination of time and cause of death.
16S rRNA sequencing can also facilitate the detection and identification of pathogenic microbes within the body tissues that could potentially be related to the cause of death, such as bacterial infections.
References:
- Hou W, Wang S, Dong H, et al. A comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan Province China using 16S rRNA gene pyrosequencing. PloS one, 2013, 8(1): e53350.
- Qian Y, Yang X, Xu S, et al. Detection of microbial 16S rRNA gene in the blood of patients with Parkinson's disease. Frontiers in aging neuroscience, 2018, 10: 156.
- Du J, Xiao K, Huang Y, et al. Seasonal and spatial diversity of microbial communities in marine sediments of the South China Sea. Antonie Van Leeuwenhoek, 2011, 100: 317-331.
- He K, Liu M, Wang Q, et al. Combined analysis of 16S rDNA sequencing and metabolomics to find biomarkers of drug-induced liver injury. Scientific Reports, 2023, 13(1): 15138.
- Morel A S, Dubourg G, Prudent E, et al. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. European Journal of Clinical Microbiology & Infectious Diseases, 2015, 34: 561-570.
- Prabhavathy G, Rajasekara M, Senthilkumar B. Identification of industrially important alkaline protease producing Bacillus subtilis by 16s rRNA sequence analysis and its applications. Int. J. Res. Pharma. Biomed. Sci, 2013, 4: 332-338.
- Hu N, Lei M, Zhao X, et al. Analysis of microbiota in Hainan Yucha during fermentation by 16S rRNA gene high‐throughput sequencing. Journal of Food Processing and Preservation, 2020, 44(7): e14523.
- Wang S, Song F, Gu H, et al. Comparative evaluation of the salivary and buccal mucosal microbiota by 16S rRNA sequencing for forensic investigations. Frontiers in Microbiology, 2022, 13: 777882.
- Speruda M, Piecuch A, Borzęcka J, et al. Microbial traces and their role in forensic science. Journal of Applied Microbiology, 2022, 132(4): 2547-2557.


 Sample Submission Guidelines
Sample Submission Guidelines Figure 1. Microbial community composition grouped by pH and temperature. (Hou et al., 2013)
Figure 1. Microbial community composition grouped by pH and temperature. (Hou et al., 2013) Figure 2. Bacterial community composition of the saliva and buccal mucosa at the phylum and genus levels. (Wang et al., 2022)
Figure 2. Bacterial community composition of the saliva and buccal mucosa at the phylum and genus levels. (Wang et al., 2022)