Ultra-Low Input RNA Sequencing: Applications, Platforms, And Advantages
RNA sequencing is a valid method for studying the transcriptome, the set of all RNA molecules transcribed under a specific developmental period or functional state in one cell or a population of cells. The transcriptome of a selected organ or tissue in an organism under a specific condition can be obtained quickly and completely via the high-throughput next-generation RNA sequencing, which covers gene expression quantification, differential gene expression analysis, variants detection, characterization of alternative splicing patterns, and small RNA profiling. RNA-seq can assist in understanding the complex biology of gene expression and regulation, exploring the unique characteristics of individual cells in tissues, and understanding the response of cell subpopulations to environmental cues. RNA-Seq also has great potential in medical research such as biomarkers profiling, druggable pathway inferring, and genetic diagnoses.
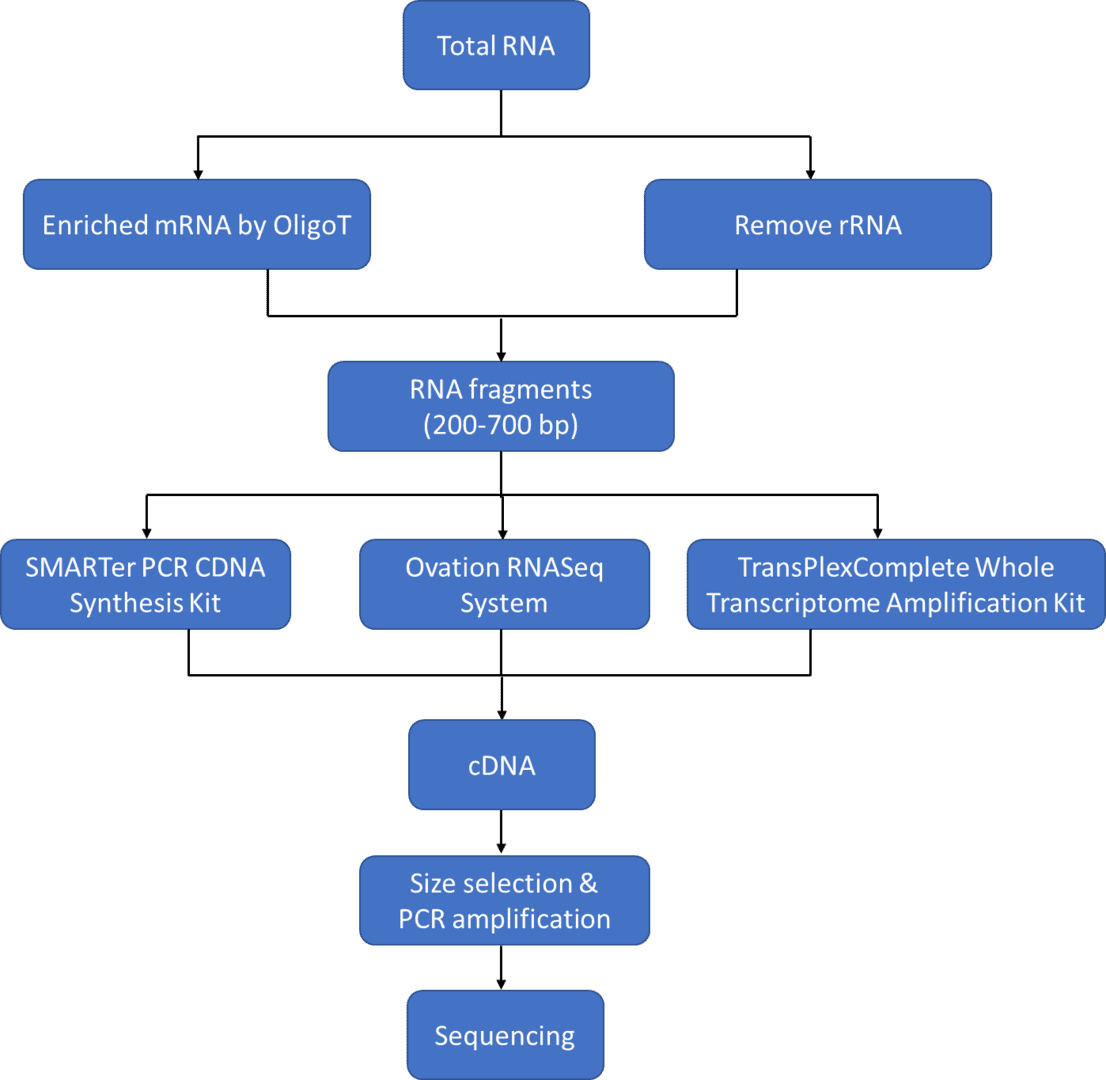
However, the strict sample requirements and the acquisition of large amounts of high-quality RNA can sometimes be demanding, making it difficult for situations such as sequencing on individual cells level or where only scarce or degraded RNA can be obtained. Fortunately, the amplification of ultra-low RNA samples can solve this problem. The recent advances in sequencing technologies empowered by next-generation sequencing (such as Illumina sequencing platforms) have facilitated the applications of RNA-Seq in multiple fields by proposing solutions to enabled RNA-seq of low-quality and/or low-quantity samples. The protocols, which can be performed by either employing full-length sequencing strategy or conducting 3' differential expression RNA sequencing using poly-A selection or rRNA depletion, are available for profiling various types of RNA including but not limited to mRNA, lncRNA, and smRNA.
 |
Ultra-low input RNA-Seq has provided a powerful alternative approach to transcriptomic studies and facilitate new discoveries with regard to tissue composition, transcriptional dynamics, and regulatory relationships between genes. Rapid technological developments at the level of cell capture, phenotyping, molecular biology, and bioinformatics point to an exciting future with numerous biological and medical applications. | |
RNA viruses, such as coronavirus, HIV, HAV, and influenza viruses, pose serious threats to human health. The development of accurate and sensitive RNA virus sequencing technologies would surely benefit public health management and promote research on antiviral methods. Combined amplification of ultra-low amounts of viral RNA with Illumina sequencing, ultra-low input RNA-Seq is capable of generating full or near full-length viral genomes from clones and clinical samples with low amounts of viral RNA and can be extended to a large number of samples, excluding the interference of host contamination. |
 | |
It is recommended that the RNA amount should be above 2 µg and the concentration should be above 50 ng/µl for regular RNA-Seq provided by most commercial platforms, while ultra-low input RNA-Seq can reduce the sample requirements remarkably to nanograms or even to picograms. If your samples are tissue cells, hundreds or dozens of cells are enough for ultra-low input RNA-Seq, and few or even single cells may as well be acceptable, which provides the opportunity to study differences between cells or cell types with an unprecedented resolution.


 Sample Submission Guidelines
Sample Submission Guidelines
