The Applications of Bioinformatics in Microbial Technology
Bioinformatics is a discipline developed on the basis of biology, mathematics and computer science. It effectively acquires and analyzes biological data such as nucleic acid sequences and protein structures, so as to conduct comprehensive and accurate biological analysis. Bioinformatics is an extremely important tool to study the microorganisms, which are widespread and highly varied. In the realm of microbiology, the organization and application of data through bioinformatics can profoundly enhance the quality of microbial research.
Introduction to Bioinformatics

Bioinformatics represents an interdisciplinary field, synthesizing concepts from computer science, mathematics, and biology. The discipline involves obtaining, processing, storing, distributing, analyzing, and interpreting biological data, drawing on computational and mathematical methodologies, and employing biological tools to elucidate and comprehend the wealth of information encapsulated within such large datasets. The aim of bioinformatics is to enrich biological data and apply computer algorithms for its analysis. Essentially, it is concerned with the analysis, storage, and exchange of vast quantities of biological data. For bioinformatics, the biological content represents the research goal, entity, and object, while information science provides the research platform, technologies, and means.
Recognized as an emerging interdisciplinary science, Bioinformatics has evolved into one of the fastest developing scientific disciplines over the past two decades. This growth is attributable not only to significant advancements in research methodologies, technological developments within Biology, particularly the continual progress in omics technology, that result in exponentially growing volumes of biological data, but also to the rapid pacing of Information Science, especially in Computer Science technology. Consequently, the storage and analysis of large-scale genomic, transcriptomic, and proteomic data sets are no longer computing challenges. The advent of topical computer algorithms, such as Machine Learning and Artificial Intelligence, allows for a deeper understanding and extraction of biological data, furthering our knowledge about the language of life with the aid of computer technology. Bioinformatics comprises pivotal domains such as biological databases, sequence alignment, gene and promoter prediction, molecular phylogenetics, structural bioinformatics, genomics, and proteomics.
The applications of bioinformatics in microbial research are manifold, offering critical support in domains such as species classification of microorganisms, gene function and metabolic pathways, regulatory patterns of gene expression, and the development of drugs and antibiotics. With the continuous advancement of bioinformatics technology, it is anticipated that this technique will maintain its crucial role in the field of microbial research, thereby catalyzing the progression and advancements within microbiology as a discipline.
Services you may interested in
Bioinformatics in Microbial Identification
The identification of genetic diversity of a large number of microorganisms is a data-intensive project, and bioinformatics can greatly improve its efficiency. Through protein sequence, DNA sequence, and protein structure information, bioinformatics technology can provide researchers with a precise and rapid means to profile microbes. For example, the single-molecule sequencing provided by PacBio SMRT can utilize fluorescent signals for microorganism sequencing and realize the synchronization of synthesis and sequencing. The application of sequencing technologies, such as Sanger sequencing and Next-Generation Sequencing (NGS), enables the acquisition of microbial genomic sequences. Leveraging bioinformatics tools for sequence alignment and species identification, we can determine the taxonomic position of microorganisms with precision, thus contributing to research on their phylogenetic relationships. Simultaneously, by contrasting and interrogating differences between various microbial genomes, we can elucidate the mechanisms underlying microbial evolution and environmental adaptation processes.
The work of Reller et al. is a prime example. These researchers identified a new species in clinical medicine through sequencing technologies, followed by bioinformatics analysis. The procedure not only enhanced our ability to detect emerging pathogens but also refined taxonomic relationships. It might further deepen our understanding of the mechanisms by which microbes induce disease. After identifying microbes, increasing amounts of information are being uploaded to relevant databases like NCBI, facilitating extensive analysis and research on environmental microbial diversity. This ongoing development propels the advancement of microbiomics.
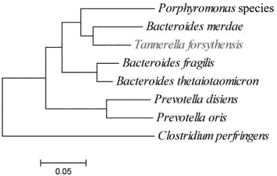 Figure 1. Reporting of novel species using phylogenetic trees. (Reller et al., 2007)
Figure 1. Reporting of novel species using phylogenetic trees. (Reller et al., 2007)
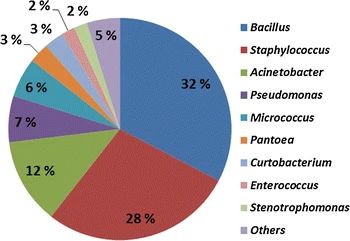 Figure 2. Pie chart of microbial diversity. (Fykse et al., 2015)
Figure 2. Pie chart of microbial diversity. (Fykse et al., 2015)
Bioinformatics in Microbial Traceability Analysis
The phylogenetic relationship of microorganisms is relatively complex, so it is challenging to conduct the tracing analysis. By bioinformatics analysis, scientists can readily build evolutionary trees and describe the evolutionary relationships among species and molecules, fulfilling various needs in pharmaceutics, diagnostics, food industry, environmental protection, etc.
Bioinformatics has valuable applications in tracing origins. Through sequencing the entire genome of a microbe, more comprehensive microbial genetic information can be obtained, which is crucial in tracking the source and variation of the microbe. By analyzing the whole genome of a microbial community, the interrelationships and functions among various microbes can be revealed, providing a more in-depth understanding for the monitoring and research of microbes. Furthermore, mathematical models can be established using bioinformatics methods to predict the dispersion routes and ecological behaviors of microbes, offering valuable insights for the tracking and control of microbes.
The intricate phylogenetic relationships of microbes are a challenge to unravel. A key application of bioinformatics in this domain is the construction of phylogenetic trees to depict evolutionary relationships between species or molecules. Notably, through their usage of virus screening methods at the molecular level in August 2005, Swedish scientists discovered a novel human bocavirus (HBoV) in the respiratory secretions of children and completed full-genome sequencing of this virus. By August 2006, the first case of HBoV was also detected in China. This virus, which is difficult to differentiate from other respiratory viruses, has attracted extensive attention from numerous experts and scholars. For example, Zhang and his colleagues utilized phylogenetic analysis to evaluate the microbial communities in Chinese Fen flavor Daqu and assess environmental impact factors. Tracing the origins of microbes in Daqu contributes to articulating the influence of external microbes on the Daqu microbial environment, thereby preserving its stability.
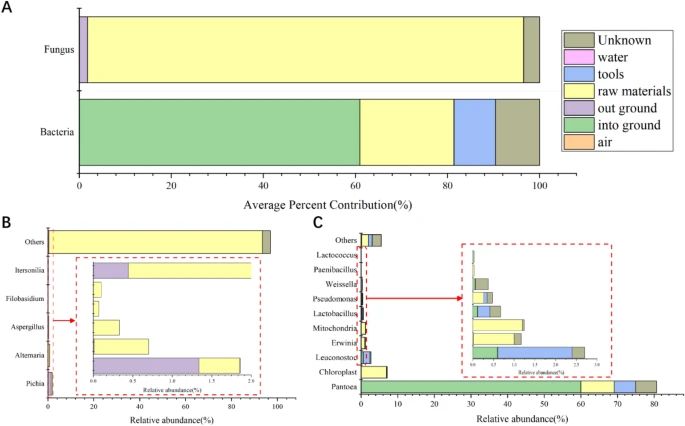 Figure 3. Microbial traceability analysis of NDaqu to determine the contribution rate of different sources. (Zhang et al., 2023)
Figure 3. Microbial traceability analysis of NDaqu to determine the contribution rate of different sources. (Zhang et al., 2023)
The FDA has established the United States Open Genome Sequencing Network, comprised of state, federal, international, and commercial partners. The GenomeTrakr network represents the first distributed genomic surveillance shield for characterizing and tracing the origins of foodborne outbreak pathogens. The GenomeTrakr network is spearheading investigations and regulatory actions for foodborne disease outbreaks, facilitating more accurate and expedited recalls of contaminated food and enhancing the monitoring of preventive control measures in food production environments. The expanded network will contribute to the establishment of an international rapid monitoring system for pathogen tracking, crucial for supporting effective public health responses to bacterial outbreaks.
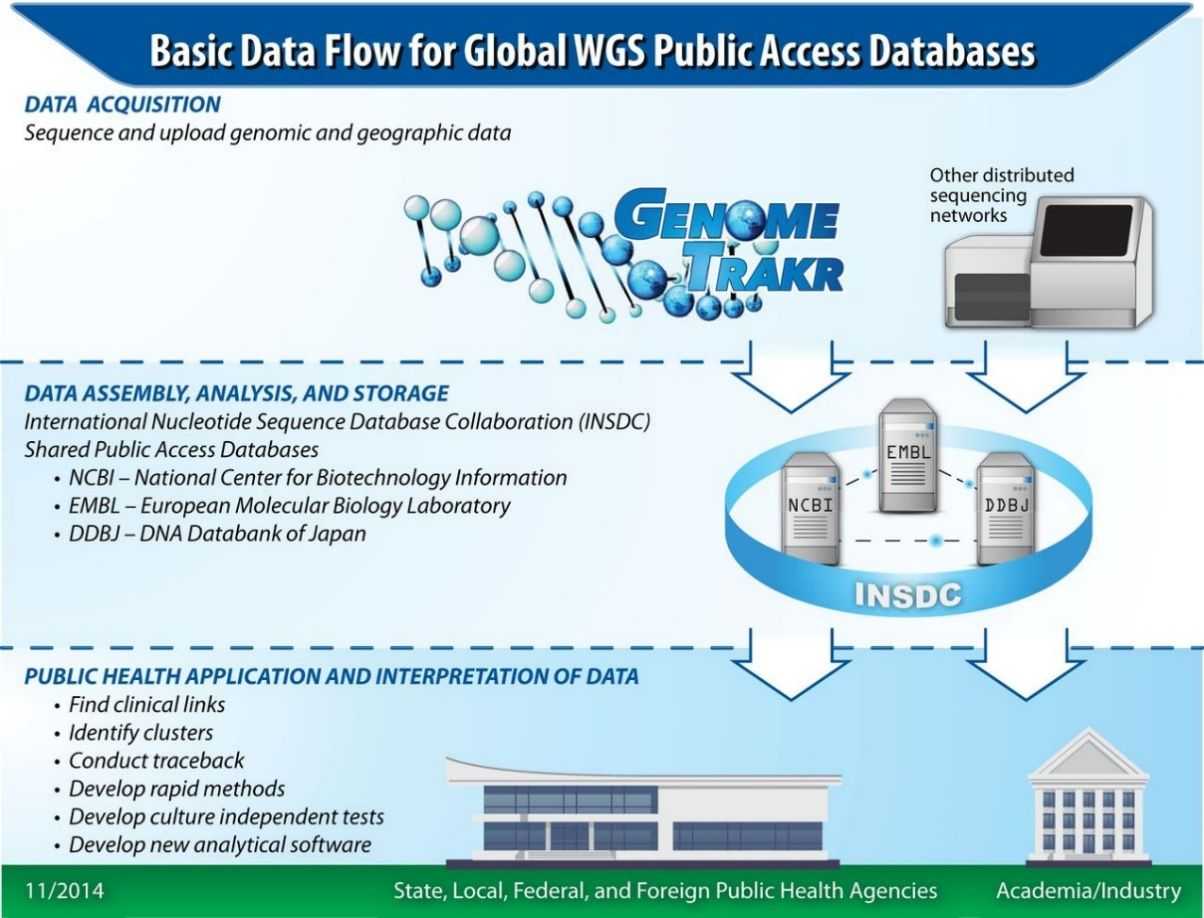 Figure 4. Data flow for the GenomeTrakr database and network. (Allard et al., 2016)
Figure 4. Data flow for the GenomeTrakr database and network. (Allard et al., 2016)
Bioinformatics in Pathogenic Gene Detection
By conducting genome-wide homology analysis, the pathogenic microorganisms and their virulence genes can be identified efficiently and extensively. For example, through the establishment of a genome barcode database by high-throughput sequencing and bioinformatics technology, the annotation of massive sequence data of microbial genomes can be achieved, which is of great practical significance for the prevention, diagnosis, and treatment of diseases.
Presently, the field of microbiology is transitioning its focus from individual gene studies to research involving multiple genes or even inter-genomic interactions, with increasing specificity. Comparative homology of entire genomic sequences enables the identification of specific antigens at various levels, such as genus, group, species, type, or subtype of pathogenic bacteria. Virulence islands, exogenous DNA segments integrated into the bacterial genome, are of particular interest. These unique segments are found in pathogenic bacterial strains that confer toxicity, but are absent in adjoining non-pathogenic strains.
Owing to the inherent characteristics of microbes, bioinformatics has proved instrumental in furthering research into the species composition, cell population, and ecological function of different microbial taxa in natural environments. It also provides direction for pharmaceutical companies and other entities in their pursuit of microbial resource development and exploitation, as well as strain isolation and screening. Additionally, bioinformatics delivers vital information concerning the composition, distribution, quantity, and other types of data related to microbial populations under varying environmental conditions.
Bioinformatics In the Development of Novel Vaccines
Bioinformatics, since its inception, has been playing an integral role in revolutionary advancements of microbial research. One stark example reflects in the context of the vaccine development process for the Neisseria meningitidis Group B, the first instance where microbial genomic sequence formed the bedrock of a new vaccine. This whole-genome-oriented approach was proven to be an effective strategy for the development of a wide range of pathogenic microorganism's vaccines. As the field of microbial genomics and bioinformatics continues to evolve, it is revolutionizing the landscape of vaccine investigation. In the contemporary setting of novel vaccine discovery, the role of bioinformatics stretches from the identification of vaccine targets to the prediction of antigenicity, and onto the design and optimization of the vaccines. The application of bioinformatics technology allows an acceleration in the vaccine development process, enhancing their safety and efficacy markers. This forms a robust tool for the prevention and containment of infectious diseases, heralding a transformative epoch in microbial research.
Likewise, Ge and his colleagues carried out an open reading frame (ORF) analysis and extracellular protein prediction of Streptococcus sanguinis, one of the many bacteria involved in dental plaque formation. While usually harmless to humans, this bacterium can potentially cause lethal infective endocarditis if it enters the bloodstream. Their bioinformatics-driven analysis yielded 43 candidate protein antigens which, after animal model experiments and affinity chromatography purification, resulted in 9 distinct antibodies. Further tests including antisera assays, competitive enzyme-linked immunosorbent assays, and fluorescence-activated cell sorting (FACS) showed strong reactivity between the purified antibodies and both the nine proteins and S. sanguinis, indicating that these proteins are exposed on the surface of the bacterium. The research findings suggest that these nine extracellular proteins could serve as reference antigens for the development of new vaccines.
Bioinformatics in Drug Production
Microorganisms are diverse as well as their metabolites, which offers the foundation for the development and research of new drugs. For example, in the field of antibiotic research, bioinformatics is widely used for analyzing microbial gene sequences, developing approaches for microbial synthesis, and enzyme engineering.
The myriad diversity and metabolic capabilities of microorganisms thriving in nature propel the development of novel microbial entities. In the realm of life sciences, the adoption of innovative microbial agents through molecular biology techniques can offer robust support for drug discovery. Coordinated biosynthesis, involving pathways orchestrated by encoded participation, exploits the interchangeability among enzyme genes, yielding heterozygous gene hybrids in the investigation of microbial secondary metabolites. Consequently, this fosters the production of multiple natural compounds. Many researchers have delved into genomic studies utilizing antibiotics like erythromycin to explore the synthesis of composite organisms. Through such studies, pivotal genes implicated in pathogenesis have been unearthed, delineating their roles in substance synthesis, metabolic breakdown, and host tissue interactions. Engaging in bioinformatics research enriches the repository of information resources, potentially unveiling new drug targets and contributing to antigenicity studies.
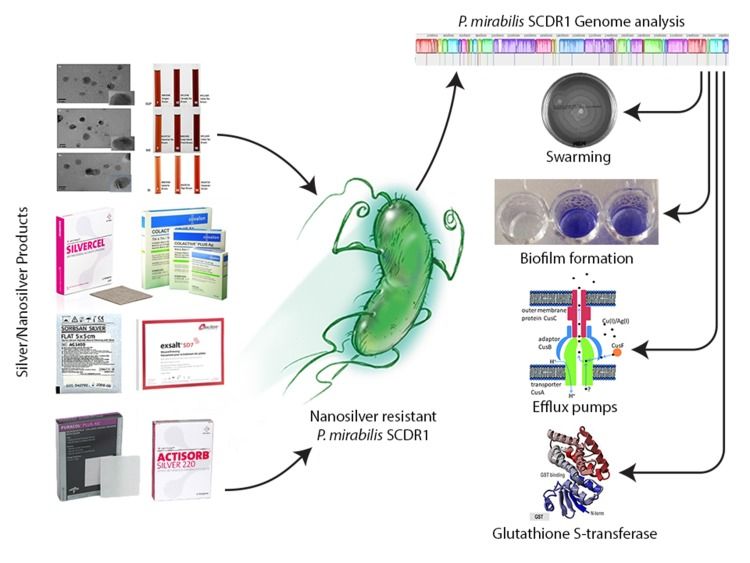 Figure 5. Resistome analysis of the first nanosilver resistance bacterium using the bioinformatics tools for identifying and combating anti-microbial resistance. (Saeb et al., 2018)
Figure 5. Resistome analysis of the first nanosilver resistance bacterium using the bioinformatics tools for identifying and combating anti-microbial resistance. (Saeb et al., 2018)
Generally speaking, features of bioinformatics include searching (data collection and screening), processing (editing, sorting, management, and presenting), and utilizing (calculation and simulation) of biological data. Researchers can use mature bioinformatics tools (specialized websites, software), as well as statistical methods and the related for many areas of study. Bioinformatics can widely cover the requirement for gene alignment, gene detection and interpretation, structure comparison, phylogenic analysis, protein structure prediction, and more, which enormously facilitates the research in microbial technology.
References:
- Bansal A K. Bioinformatics in microbial biotechnology–a mini review. Microbial Cell Factories, 2005, 4: 1-11.
- Dahiya B P. Bioinformatics impacts on medicine, microbial genome and agriculture. Journal of Pharmacognosy and Phytochemistry, 2017, 6(4): 1938-1942.
- Hiraoka S, Yang C, Iwasaki W. Metagenomics and bioinformatics in microbial ecology: current status and beyond. Microbes and environments, 2016, 31(3): 204-212.
- Kumar A, Chordia N. Role of bioinformatics in biotechnology. Res Rev Biosci, 2017, 12(1): 116.
- Hiraoka S, Yang C, Iwasaki W. Metagenomics and bioinformatics in microbial ecology: current status and beyond. Microbes and environments, 2016, 31(3): 204-212.
- Cheba B A. Review on Microbial Bioinformatics: Novel and Promoting Trend for Microbiomics Research and Applications//International Conference Interdisciplinarity in Engineering. Cham: Springer International Publishing, 2021: 718-729.
- Saeb A T M. Current Bioinformatics resources in combating infectious diseases. Bioinformation, 2018, 14(1): 31.
- Reller L B, Weinstein M P, Petti C A. Detection and identification of microorganisms by gene amplification and sequencing. Clinical infectious diseases, 2007, 44(8): 1108-1114.
- Fykse E M, Tjärnhage T, Humppi T, et al. Identification of airborne bacteria by 16S rDNA sequencing, MALDI-TOF MS and the MIDI microbial identification system. Aerobiologia, 2015, 31: 271-281.
- Zhang Y, Xu C, Xing G, et al. Evaluation of microbial communities of Chinese Feng-flavor Daqu with effects of environmental factors using traceability analysis. Scientific Reports, 2023, 13(1): 7657.
- Allard M W, Strain E, Melka D, et al. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. Journal of clinical microbiology, 2016, 54(8): 1975-1983.
- Ge X, Kitten T, Munro C L, et al. Pooled protein immunization for identification of cell surface antigens in Streptococcus sanguinis. PLoS One, 2010, 5(7): e11666.


 Sample Submission Guidelines
Sample Submission Guidelines
