The Application of Nanopore Sequencing in Tuberculosis Research
Nanopore sequencing technology was developed by Oxford Nanopore Technologies Ltd, and is the most powerful method for rapid generation of long-read sequences. Due to the unrestricted read length and system portability, Nanopore sequencing opens the possibility of competitive cost real-time sequencing and data analysis. One of the main applications in medical scenarios is the rapid diagnosis of infectious diseases, which has the potential to enhance our ability to diagnose, interrogate, and track infectious diseases, so as to timely and accurately manage patients and make treatment decisions.
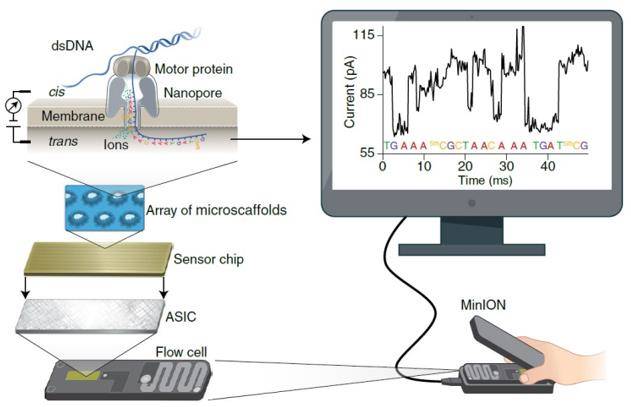 The principle of nanopore sequencing
The principle of nanopore sequencing
Infectious diseases are the main driving force of global incidence rate and mortality and an enormous public health burden and a growing threat to human health worldwide. Emerging classical recurrent pathogens or pathogens with drug-resistance characteristics challenge our ability to diagnose and control infectious diseases. The treatment of malaria, tuberculosis and human immunodeficiency virus infections is particularly challenging, as demonstrated by the continued spread and high mortality rates of these diseases.
Mycobacterium tuberculosis is a pathogen that imperil public health and cause tuberculosis. Nanopore sequencing can also identify methylation status, which is important because epigenetic modifications in M. tuberculosis have been associated with drug resistance, virulence, and regulation of gene expression profiles. Recently, the reduction of error rate, the update of flow cytometry, the reduction of the amount of DNA required for input, and the faster library preparation scheme have aroused renewed interest in its application in tuberculosis research and clinical application. At present, it has solved many problems in the research and clinical fields of tuberculosis.
The advantages of Nanopore sequencing in tuberculosis research
(1) The construction of the library is simple, and the construction of the library takes only 10 minutes at the fastest.
(2) Due to the low hardware investment, simple operation process, compatibility with automatic library preparation, and the continuous improvement of sequencing accuracy of the Nanopore sequencing equipment, it will also play an increasingly important role in the detection of M. tuberculosis, especially the related detection of drug resistance, which has greatly accelerated the clinical process of the application of the Nanopore sequencing technology in the detection of M. tuberculosis.
(3) The sequencing speed is fast, and the sequencing and data analysis can be completed within 1 hour at the fastest.
(4) The key strengths of Nanopore sequencing are that they have competitive per-sample sequencing cost when multiplexing, the possibility of bias-free PCR library preparation, cold-chain-free sequencing reagents, fast turnaround time, use of long reads to resolve complex genomic loci, and the ability to investigate methylation status.
(5) Nanopore sequencing has enabled many biomedical studies by providing ultralong reads from single DNA/RNA molecules in real time. Using nanopore sequencing, a single molecule of DNA or RNA can be sequenced without the need for PCR amplification or chemical labeling of the sample.
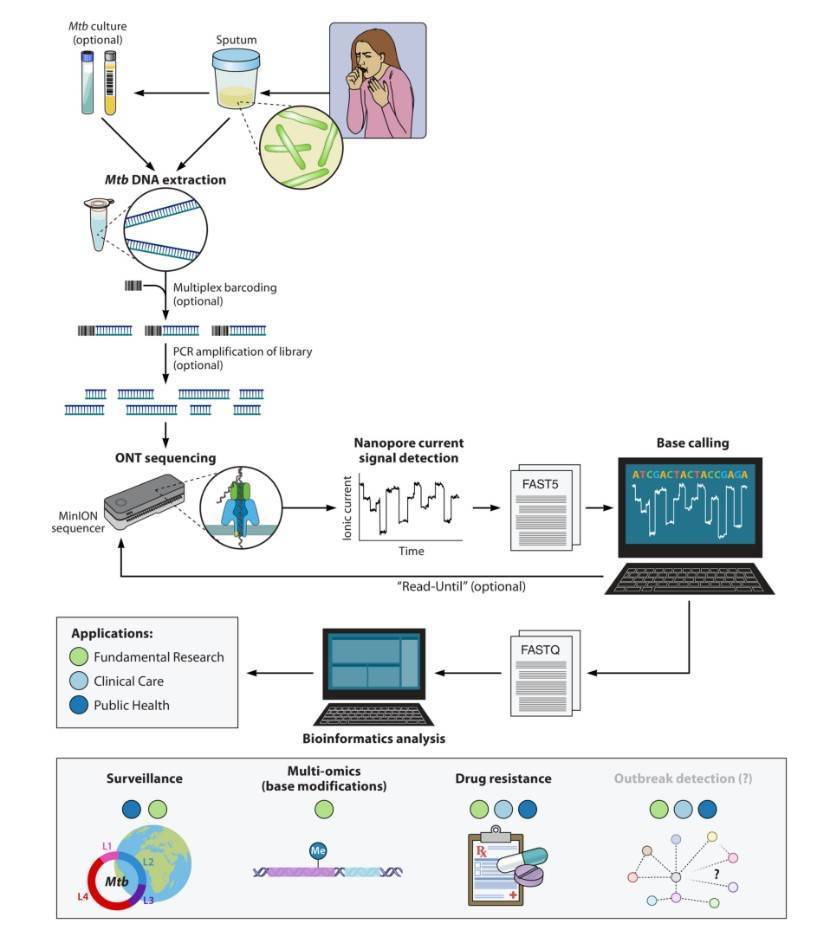 The Mycobacterium tuberculosis sequencing approach using Oxford Nanopore Technologies sequencing platform
The Mycobacterium tuberculosis sequencing approach using Oxford Nanopore Technologies sequencing platform
Future application value of Nanopore sequencing in tuberculosis research
The low capital cost and portability of ONT hardware, the simplification and automation of sample and library preparation steps when using VolTRAX, and the continuous improvement of sequencing accuracy indicate that ONT is valuable in mycobacterium research laboratories, especially for the detection of drug resistance. What's more exciting is that Nanopore sequencing may accelerate researchers' ability to sequence directly from sputum samples and may expand the research applications in M. tuberculosis sequencing beyond what is possible using short-read sequence analysis workflows by including epigenetics and investigations of the role of repetitive elements and complex regions of the M. tuberculosis genome. Nanopore sequencing has enhanced our ability to study complex M. tuberculosis samples that has allowed to address several previously unsolved issues.
Due to its natural advantages in epigenetic detection, genome repeat element and other special sequence sequencing, as the research results of Nanopore sequencing technology applied to the detection of M. tuberculosis increase year by year, it is believed that Nanopore sequencing technology will gradually enter the clinical detection field related to tuberculosis, making greater contributions to more public health undertakings in the future.
References:
- Zhang, Lu Lu et al. "Application of Nanopore Sequencing Technology in the Clinical Diagnosis of Infectious Diseases." Biomedical and environmental sciences: BES vol. 35,5 (2022): 381-392.
- Chan, Wai Sing et al. "Rapid and economical drug resistance profiling with Nanopore MinION for clinical specimens with low bacillary burden of Mycobacterium tuberculosis." BMC research notes vol. 13,1 444. 18 Sep. 2020.
- Dippenaar A, Goossens SN, Grobbelaar M, et al. Nanopore Sequencing for Mycobacterium tuberculosis: a Critical Review of the Literature, New Developments, and Future Opportunities. Journal of Clinical Microbiology. 2022 Jan;60(1): e0064621.
- Wang, Yunhao et al. "Nanopore sequencing technology, bioinformatics and applications." Nature biotechnology vol. 39,11 (2021): 1348-1365.


 Sample Submission Guidelines
Sample Submission Guidelines
