Single-cell RNA Sequencing Enables Tumor Research
Single cell RNA sequencing (scRNA-seq) technology has revolutionized our understanding of cellular heterogeneity by providing in-depth transcriptome information at the single cell level, which has been widely used in many fields. With the development of sequencing technology, cell separation and whole genome amplification technology, the single cell sequencing technology for sequencing genomes at the single cell level is gradually emerging. On the one hand, it solves the problem that micro samples cannot be detected, and on the other hand, it better analyzes the heterogeneity of genetic variation between different cells.
The single cell sequencing process includes: isolation and preparation of single cells, extraction and amplification of genetic material, and high-throughput sequencing of genetic material. With the arrival of the era of precision therapy, it has been applied to a series of biomedical research and clinical trials, such as tumor detection, embryonic development, immune cell therapy and stem cell differentiation, and is becoming the focus of life science research.
Single-cell analyses reveal suppressive tumor microenvironment of human colorectal cancer
scRNA-seq has been widely used to investigate cellular heterogeneity in almost all cancer types with substantial advances in available experimental techniques and bioinformatics pipelines in recent years. Tumor microenvironment (TME) influences both tumor progress and response to immunotherapy. Single-cell RNA sequencing (scRNA-seq) provides unprecedented detailed characterization of transcriptomes of cell diversity and heterogeneity thereby allowing comprehensive assessment of the complexity of tumor microenvironment. Researchers generated 34,037 single-cell transcriptomes and chromatin accessibility landscape of 6525 single cells by sampling from 12 CRC patients. Using single-cell ATAC-seq and profiling epigenetic landscape and inferring transcription factor motifs for further explored the underlying molecular mechanisms. This study provides a rich resource for further study of the impact of immune cells and translational research for human colorectal cancer. Most importantly, it reveals that immune cells and nonimmune cells together constitute the complex tumor microenvironment components of CRC, which provides valuable reference data for the treatment of human colorectal cancer as a new target of immunotherapy.
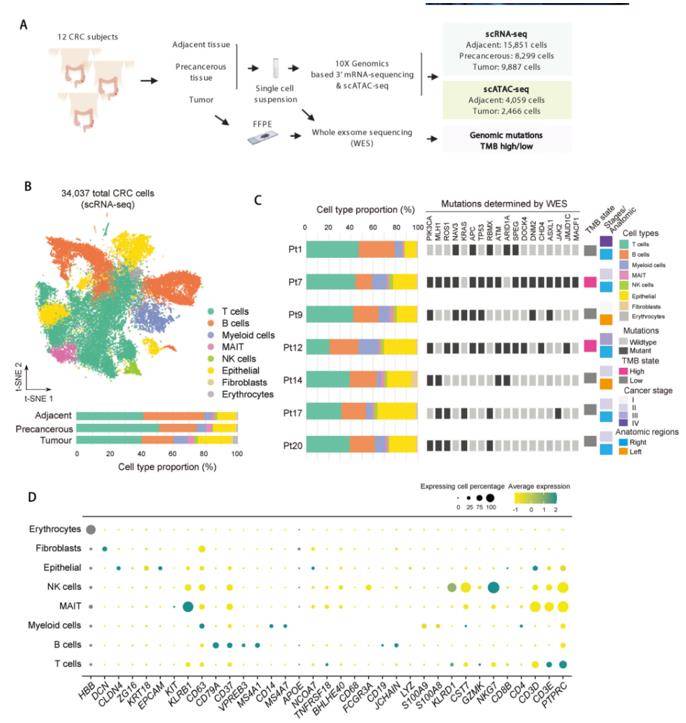 Single-cell RNA-seq profiles of the human CRC
Single-cell RNA-seq profiles of the human CRC
scRNA-seq for analyzing the malignant phenotype and development trajectory
Childhood malignancies likely arise from precursor cells during embryonic or early postnatal development. Neuroblastoma (NB) has been confirmed the most common extracranial solid tumor occurring in childhood. Recent advances in single-cell research have strengthened the associations between tumorigenesis of several pediatric cancers and abnormal development of relevant fetal cells.
At present, the recognition of phenotypic diversity of NB at single-cell resolution is still lacking. Single-cell RNA sequencing (scRNA-seq) was performed on early human embryos, fetal adrenal glands, and primary adrenal NB tumors aimed to clarify the phenotypes of NB tumor cells by relating them to their putative developmental origins. The majority of adrenal NB tumor cells are transcriptionally similar to noradrenergic chromaffin cells. Malignant states also mimic the proliferation/differentiation status of chromaffin cells during normal development. It is the first time to analyze the single cell map of neuroblastoma in children by using single-cell RNA sequencing, which provides a new perspective for people to deeply understand the pathogenesis of neuroblastoma.
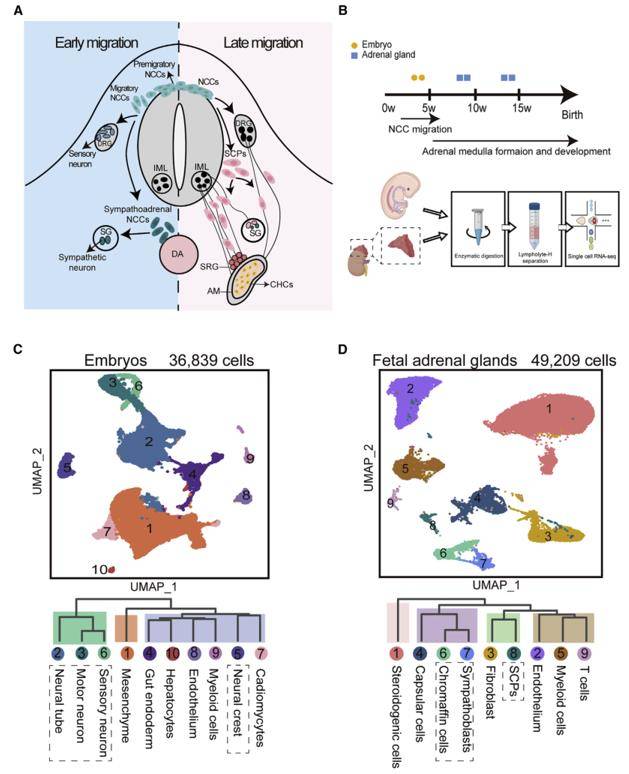 Single-Cell Transcriptomic Profiling of Early Human Embryos and Fetal Adrenal Glands
Single-Cell Transcriptomic Profiling of Early Human Embryos and Fetal Adrenal Glands
Single cell sequencing technology is universal comprehensive and multi-level, and all individual eukaryotic cells with cells can be studied with this technology. That is to say, it can be used to study almost any species we know. Additionally, single cell sequencing can not only measure the gene expression level more accurately, but also detect a small amount of gene expression or rare non coding RNA. With the arrival of the era of precision therapy, the future single cell technology will help human scientific research and clinical application, especially in the fields of tumor, microbiology, neuroscience, immunology and so on. We firmly believe that single cell sequencing technology will definitely promote precision medicine to a wider space, and more patients will be able to enjoy specific and efficient therapy that can be affordable and improve the treatment efficiency and experience of the treatment process. On the stage of human health, it will be more brilliant.
References:
- Dong R, Yang R, Zhan Y, et al. Single-Cell Characterization of Malignant Phenotypes and Developmental Trajectories of Adrenal Neuroblastoma. Cancer Cell. 2020;38(5):716-733.e6.
- Mei Y, Xiao W, Hu H, et al. Single-cell analyses reveal suppressive tumor microenvironment of human colorectal cancer. Clin Transl Med. 2021; 11(6): e422.


 Sample Submission Guidelines
Sample Submission Guidelines
