Genomics and Transcriptomics Reveal Luciferase Clusters in Fungi
Fungal genome sequencing encompasses several key techniques, including fungal genome scanning, fine mapping, and resequencing. This powerful approach enables the prediction of crucial genes and proteins, facilitating a deeper understanding of their functions and underlying mechanisms. Fungal genome sequencing has supplanted conventional methods, emerging as an indispensable tool for investigating the genetic mechanisms behind bacterial evolution and essential functional genes.
The genus Mycena comprises over 600 species known for their distinctive umbrella-shaped caps, and their habitat spans the globe. Notably, Mycena fungi exhibit a captivating trait – bioluminescence. Of the 81 luminescent fungi species identified, 68 belong to Mycena, a phenomenon that has piqued the interest of researchers. It has been established that luciferase plays a pivotal role in fungal luminescence and is highly conserved among fungi. Nevertheless, the evolutionary origin of luciferase in fungi and the genes associated with luminescence remain shrouded in mystery. In response to this, researchers have developed a model to shed light on the evolution of fungal bioluminescence. They achieved this by creating high-quality reference genomes for five Mycena species, leveraging comparative genomics and transcriptomics data.
Genome Assembly and Annotation: Transposable Element Interactions and DNA Methylation
Utilizing ONT sequencing and next-generation sequencing, the researchers successfully constructed genomes for five species within the compact mushroom genus, Mycena. These genomes exhibited a size range spanning from 50.9 to 167.2 megabases, with Contig N50 values ranging from 2.2 to 5.5 megabases. The annotation process yielded a total of 13,940 to 26,334 protein-coding genes, while the genome completeness, as evaluated using BUSCO metrics, consistently ranged between 92.1% and 95.3%. In summary, the researchers have established five high-quality reference genomes for the Mycena species.
Similar to other fungal genomes, the variation in the sizes of these five selected Mycena genomes can be attributed to the content of repetitive sequences. Notably, the smallest genome, M. chlorophos, contained only 11.7% repetitive sequences, whereas the larger genomes, M. sanguinolenta and M. venus, exhibited a substantial 35.7% to 39% of repetitive sequences. Furthermore, these genomes of the Mycena species demonstrated typical fungal characteristics, characterized by low repetitive sequence content and high genome density.
Additionally, the research uncovered an intriguing relationship between genome size and methylation levels in M. venus. It was observed that the expansion of transposable element (TE) sequences within the Mycena species correlated with an increase in repetitive sequences and a concurrent reduction in methylation levels. This phenomenon ultimately led to the larger genome sizes observed.
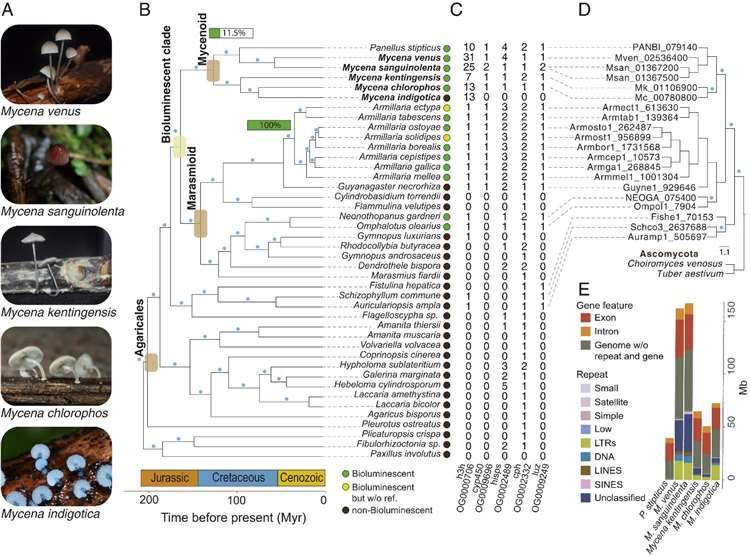 Phylogenomic analysis of Mycena and related fungi. (Ke et al., 2020)
Phylogenomic analysis of Mycena and related fungi. (Ke et al., 2020)
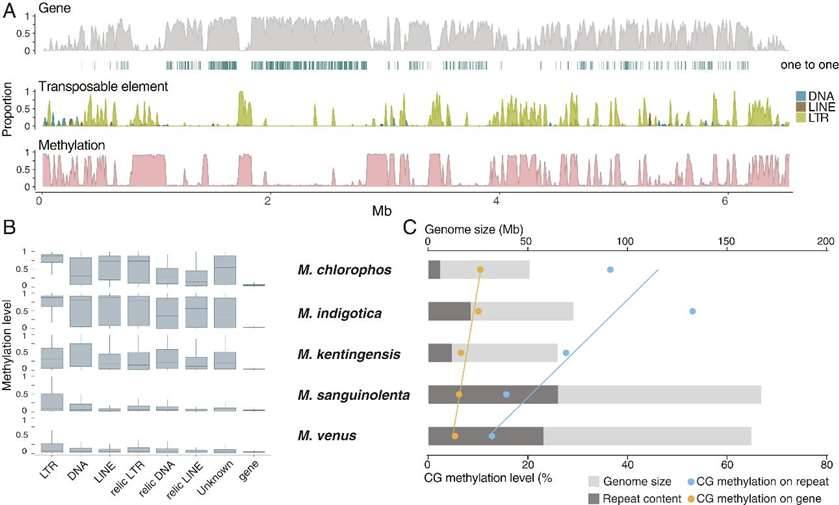 Distribution of Mycena genome features. (Ke et al., 2020)
Distribution of Mycena genome features. (Ke et al., 2020)
The Evolution of Luciferase: A Key Enzyme Driving Fungal Luminescence
Phylogenetic trees, constructed through various methodologies, strongly suggest that the origin of luciferase can be traced back to the Late Jurassic period, approximately 160 million years ago. However, an intriguing question emerges: why do luminescent fungi primarily cluster within the genera Mycena and Marasmioid, while their non-luminescent counterparts within these same genera seem to have lost the essential luciferase gene cluster? This peculiarity prompted the researchers to delve into the intricate evolutionary history of luciferase.
The research findings shed light on the fact that the luciferase of the ancestral species within luminescent fungi, encompassing key gene clusters like luz, h3h, cyp450, and hisps, were situated together on the same chromosome alongside the cph enzyme. However, over time, a distinct evolutionary trajectory unfolded within the Mycena and Marasmioid species.
Notably, the luciferase of luminescent fungi in the Marasmioid clade was confined to genomic regions where evolutionary changes occurred at a slower pace, devoid of chromosomal rearrangements. This environment allowed for the preservation of consistent luciferase characteristics among this group. On the contrary, the luciferase within the Mycena species luminescent fungi underwent multiple transposable element-mediated chromosomal rearrangement events, resulting in notable divergence among their luciferase sequences.
Moreover, the non-luminous fungi within these genera displayed a complete loss of the luciferase gene, distinguishing them from their bioluminescent counterparts.
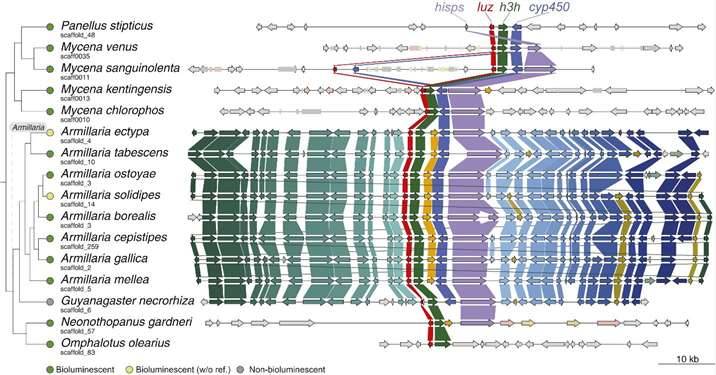 Synteny around the luciferase cluster among bioluminescent fungi. (Ke et al., 2020)
Synteny around the luciferase cluster among bioluminescent fungi. (Ke et al., 2020)
Identifying Genes Associated with Fungal Luminescence
In the pursuit of accurately pinpointing the genes responsible for fungal luminescence, the researchers employed a two-fold transcriptome analysis approach. They conducted a differential gene expression analysis across four distinct fungi, each exhibiting varying degrees of luminescence. Additionally, the researchers compared the expression patterns of luminescence-related genes in different tissues, focusing on Mycena kentingensis as a representative example.
The findings revealed a total of 29 differentially expressed genes within the four fungi, notably including luz, h3h, and hisps genes, thereby affirming their direct involvement in the luminescence process. A Weighted Gene Co-Expression Network Analysis (WGCNA) also identified 67 co-expressed genes within M. kentingensis. This set not only encompassed the aforementioned 29 luminescence-related genes but also encompassed genes associated with cell wall remodeling. This intriguing discovery implies a possible connection between the fungal development process, cell wall remodeling, and bioluminescence.
In summary, this study underscores the dynamic regulation of the luciferase cluster during fungal development, showcasing distinct levels of expression among various lineages of bioluminescent fungi. Through a comprehensive comparative genome analysis, the researchers constructed an evolutionary model of luciferase, shedding light on its evolution across various luminescent fungi spanning over 160 million years. The transcriptome analysis further unveiled a plethora of genes linked to fungal luminescence, offering fresh perspectives for the exploration of this fascinating phenomenon.
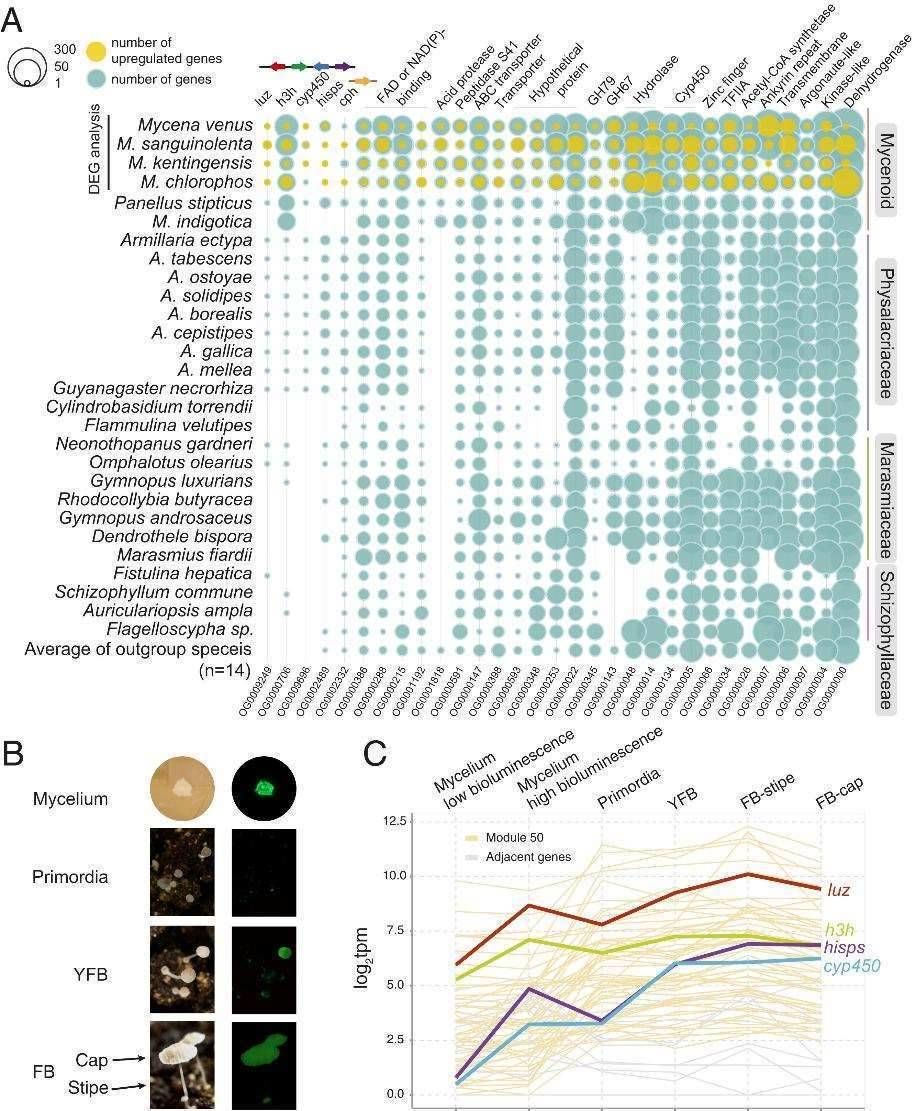 Expression analysis to identify genes involved in bioluminescence. (Ke et al., 2020)
Expression analysis to identify genes involved in bioluminescence. (Ke et al., 2020)
Reference:
- Ke, Huei-Mien, et al. "Mycena genomes resolve the evolution of fungal bioluminescence." Proceedings of the National Academy of Sciences 117.49 (2020): 31267-31277.


 Sample Submission Guidelines
Sample Submission Guidelines
