What is RNA m6A
What is The m6A Modification?
RNA methylation stands as a pivotal focus within the realm of epigenetics. With a repertoire exceeding 100 distinct modifications, notable examples include methylation, hydroxymethylation, and acetylation. Among these, m6A stands as the most prevalent methylation modification on RNA, averaging 1 to 3 m6A modifications per transcript. Ubiquitous across eukaryotes, encompassing mammals, insects, plants, yeast, and select viruses, m6A occurs not only on mRNA but also on tRNA, rRNA, small nuclear RNA, and various long non-coding RNAs. RNA methylation is poised to exert significant influence over gene expression regulation, splicing, RNA editing, stability, mRNA turnover, and degradation processes.
m6A methylation (N6-methyladenosine) denotes the process whereby RNA molecules, under the catalytic influence of the RNA methyltransferase complex (MTC), selectively add a methyl group to the N6 position of adenosine. Bases that have undergone m6A modification can undergo demethylation through the action of two enzymes, FTO and ALKBH.
You may interested in
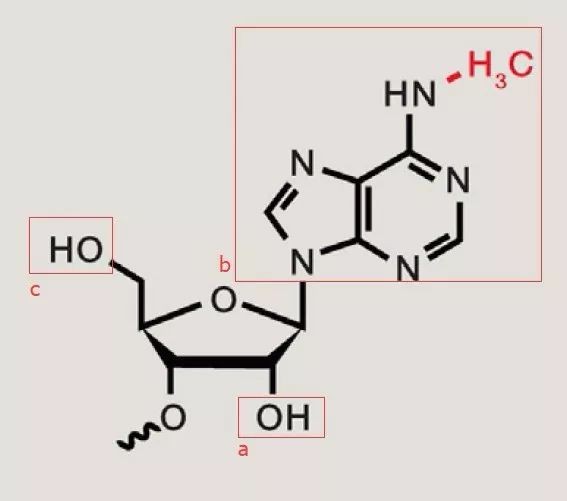 N6-methyladenosine (Roundtree et al (2017) Cell)
N6-methyladenosine (Roundtree et al (2017) Cell)
What is The Function of m6A Methylation?
| m6A modulates mRNA precursor splicing | m6A regulates RNA nuclear export |
| m6A regulates mRNA translation | m6A affects mRNA stability |
| m6A modification facilitates circular RNA translation | m6A modification alters tumor development |
| m6A regulates hematopoietic stem cell differentiation | m6A regulates spermatogenesis |
Regulation of Hematopoietic Stem Cell Differentiation by RNA Methylation Modification (m6A)
The m6A modification, mediated by METTL3, plays a crucial role in various biological processes and individual development across eukaryotes, including circadian rhythms, DNA damage response, regulation of stem cell self-renewal and pluripotency, maternal-to-zygotic transition, Drosophila neuroregulation and sex determination, and early mouse embryonic development. The discovery of RNA methyltransferases, binding proteins, and their involvement in RNA metabolism and processing underscores the significant biological functions associated with RNA methylation modification.
In vertebrates, hematopoietic stem cells originate from specialized hemogenic endothelium through the endothelial-to-hematopoietic transition (EHT) process during embryonic development in the aorta-gonad-mesonephros region. Subsequently, they migrate to and proliferate within hematopoietic tissues, then migrate to the thymus for development into lymphoid cells, and ultimately migrate to the bone marrow (in mice and humans) or kidney marrow (in zebrafish) to sustain lifelong hematopoiesis. Utilizing MeRIP-seq and m6A single-nucleotide resolution miCLIP-seq sequencing techniques with m6A antibodies, a detailed map of m6A modification during zebrafish embryonic development has been constructed. It was discovered that m6A selectively modifies key genes such as notch1a during the endothelial-to-hematopoietic transition, recruiting the binding protein YTHDF2 for mRNA degradation, thereby inhibiting the Notch signaling pathway and promoting the orderly progression of the endothelial-to-hematopoietic transition, facilitating the conversion of endothelial cells into hematopoietic stem/progenitor cells (HSPCs). In zebrafish embryos with mettl3 deficiency, the level of m6A modification significantly decreased, and notch1a could not be degraded normally, leading to sustained activation of the Notch signaling pathway and ultimately resulting in failure of endothelial cells to convert into hematopoietic stem/progenitor cells. Moreover, Mettl3 knockout mice embryos exhibited similar phenotypes. These results elucidate the regulatory mechanism of mRNA m6A methylation modification in determining the fate of vertebrate hematopoietic stem cells, highlighting the critical role of RNA epigenetic modification in hematopoietic development.
RNA Methylation Modification (m6A) Regulates Spermatogenesis
The biological function of m6A modification mediated by Mettl3 in mammalian tissue development remains elusive due to the lethality of Mettl3 knockout in early mouse embryos. Here, we established a mouse model with specific Mettl3 deletion in germ cells (Vasa-Cre), termed Mettl3 conditional knockout (Mettl3CKO), utilizing homologous recombination facilitated by the CRISPR/Cas9 system and the Cre-loxP system. Through this model, we systematically investigated the crucial role and regulatory mechanism of METTL3-mediated m6A modification in mouse spermatogenesis. Histological examination via H&E (hematoxylin and eosin) staining, immunofluorescence staining, and chromosome spread analysis revealed that Mettl3 deletion suppressed the differentiation of mouse spermatogonial stem cells and the initiation of meiosis, ultimately leading to male infertility. RNA-seq, m6AmiCLIP-seq, and bioinformatics analysis demonstrated that Mettl3 deficiency resulted in decreased m6A levels, causing aberrant RNA alternative splicing of genes related to spermatogonial stem cell maintenance and differentiation, cell cycle, and meiosis, as well as overall disruption of gene expression in germ cells, thereby impeding spermatogenesis. These findings underscore the crucial biological function of RNA methyltransferase METTL3-mediated m6A modification in mouse spermatogenesis.
Specific knockout of Mettl3 or Mettl14 in mouse germ cells using Vasa-Cre led to decreased m6A levels, disrupting the translation function of transcripts associated with spermatogonial stem cell proliferation and differentiation, ultimately depleting spermatogonial stem cells. Simultaneous knockout of Mettl3 and Mettl14 using Stra8-GFPCre disrupted the translation of haploid-specific genes during spermatogenesis, resulting in impaired spermatogenesis. Moreover, male Alkbh5-/- mice exhibited phenotypes such as testicular atrophy, abnormal spermatogenesis, and impaired sperm motility. Increased numbers of primary and secondary spermatocytes were observed in seminiferous tubules at stage VII of development, accompanied by a decrease in round spermatids and mature sperm, as well as cell apoptosis. Ythdc2 knockout mice showed excessive apoptosis in early spermatocytes during meiotic prophase, leading to testicular atrophy and abnormal sperm development. The m6A-binding capability and 3′→5′ helicase activity of YTHDC2 play crucial regulatory roles in maintaining the correct expression pattern of genes related to meiosis during spermatogenesis. Collectively, these studies highlight the dynamic balance of RNA m6A methylation modification as a crucial regulatory factor in the process of gamete (sperm) development.
RNA Methylation Modification (m6A) Regulates Brain Development
The m6A modification plays an indispensable regulatory role in both the development and function of the nervous system. Deletion of the Ime4 gene in fruit flies results in viable but shorter-lived individuals displaying significant behavioral abnormalities, indicating the impact of m6A modification loss on fruit fly neurofunctionality. Specific deletion of the Mettl14 gene in the central nervous system of mice severely affects the development of the cerebral cortex. Loss of the Ythdf2 gene in mice leads to an overall increase in m6A levels, preventing crucial factors involved in neural stem cell differentiation and neurite dendrite formation from entering the normal RNA degradation pathway. Consequently, this disruption results in impaired asymmetric division of cortical neural stem cells, causing a significant loss of neural precursor cells and severely affecting neuronal differentiation, ultimately leading to slow development of the forebrain cerebral cortex in mice. Apart from the cerebral cortex, the RNA m6A methylation pattern and level in the cerebellum are particularly noteworthy. The dynamic processes of m6A methylation and demethylation span the entire postnatal development of the cerebellum, and loss of the Alkbh5 gene under conditions of low pressure and low oxygen disrupts the m6A levels of genes involved in cerebellar development regulation, accelerating RNA nuclear export and significantly delaying cerebellar development.
To investigate the impact of m6A methylation modification on central nervous system development, we specifically deleted the Mettl3 gene in the central nervous system of mice using the Cre-loxP system. Specific deletion of Mettl3 in the central nervous system resulted in severe motor dysfunction during the lactation period and led to death. Anatomical and histopathological examination revealed that the loss of m6A methylation modification caused by Mettl3 deletion severely affected the development of the cerebral cortex and cerebellum, resulting in cortical thinning and cerebellar maldevelopment. Loss of m6A methylation modification caused dysregulation of gene expression during granule neuron differentiation and maturation in the cerebellum, leading to extensive apoptosis of newborn granule neurons, highlighting the crucial role of METTL3-mediated m6A modification in mammalian central nervous system development. In addition to its impact on central nervous system development, m6A modification also participates in adult nervous system regulation. Axonal injury promotes an increase in m6A modification levels within nerve cells, thereby enhancing the translation efficiency of a series of genes, including those related to axon regeneration.
RNA Methylation Modification (m6A) and RNA Metabolism
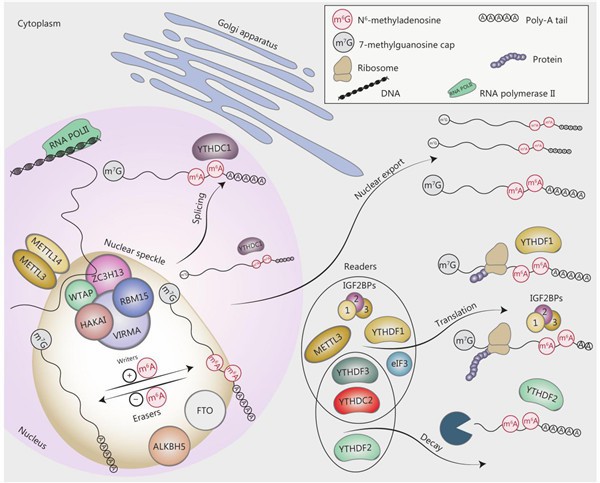
An increasing body of research suggests that m6A is intricately involved in regulating various processes of RNA metabolism, ranging from precursor mRNA processing within the nucleus, mRNA expression, to mRNA translation and degradation within the cytoplasm. Within the nucleus, m6A recruits SRSF3 through its binding protein YTHDC1, participating in the regulation of precursor mRNA splicing. It also modulates the diversity of adenine polyadenylation by regulating the selective splicing of the final exon. Moreover, the m6A modification enzyme system, including METTL3, ALKBH5, and YTHDC1, has been found to participate in regulating mRNA export processes. In the cytoplasm, the mechanisms by which m6A regulates translation are diverse. This includes pathways such as the YTHDF1-eIF3 pathway, which regulates mRNA translation efficiency, as well as processes mediated by the IGF2BP protein family. Under stress conditions, m6A participates in translation regulatory pathways that do not rely on the 5′ cap. Furthermore, m6A regulates mRNA stability through various pathways, including the recruitment of mRNA to P-bodies for degradation by YTHDF2. Recent reports have also indicated that the IGF2BP protein family maintains mRNA stability by specifically binding to m6A-modified mRNA. Additionally, m6A can regulate RNA metabolism by modulating RNA secondary structure.
The Biological Functions of RNA Methylation Modification m5C
In recent years, an increasing body of research has highlighted the crucial regulatory role of m5C modification on mRNA in RNA processing and metabolism, as well as in normal physiological processes, including RNA metabolism, stem cell differentiation, and stress responses. Studies on mouse embryonic stem cells and the brain have revealed distinct levels and distributions of m5C modification at different stages of stem cell differentiation. In Arabidopsis thaliana, m5C modification on mRNA exhibits tissue specificity and plays a vital role in plant development. Mutant plants of the m5C methyltransferase TRM4B in Arabidopsis exhibit inhibited cell division in the root apical meristem, resulting in shorter roots and heightened sensitivity to oxidative stress.
M6A Modification Mechanism
RNA methylation modification accounts for over 60% of all RNA modifications, with m6A being the most prevalent modification on mRNA and lncRNAs in higher organisms. Currently, m6A modification has been identified on microRNAs, circRNAs, rRNAs, tRNAs, and snoRNAs. The occurrence of m6A modification primarily targets adenosines within the RRACH sequence motif and its functionality is governed by the "Writer," "Eraser," and "Reader" components. The "Writer" refers to the methyltransferase complex, comprising known components such as METTL3, METTL14, WTAP, and KIAA1429. Conversely, ALKBH5 and FTO act as demethylases (Erasers) capable of reversing methylation. m6A is recognized by m6A-binding proteins (Readers), which include YTH domain proteins (such as YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2) and the heterogeneous nuclear ribonucleoprotein (HNRNP) family (HNRNPA2B1 and HNRNPC). This intricate system of writers, erasers, and readers orchestrates the regulatory landscape of m6A modification in RNA biology.
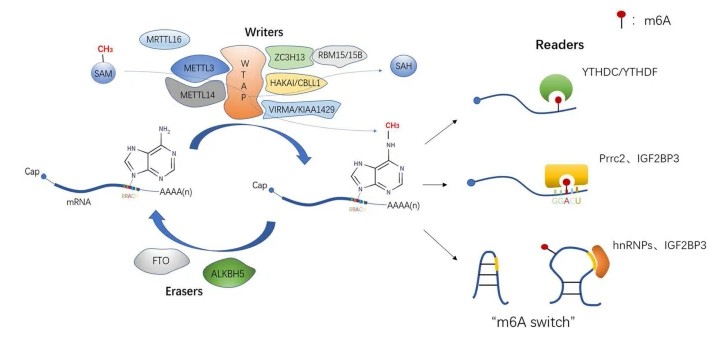 Dynamic and reversible process of m6A modification and three recognition methods of readers
Dynamic and reversible process of m6A modification and three recognition methods of readers
m6A Methyltransferases (Writers)
Methyltransferases, also known as Writers, comprise a crucial class of catalytic enzymes responsible for catalyzing m6A methylation modifications on mRNA. Writers facilitate the "writing" of methyl modifications onto RNA, thereby mediating the process of RNA methylation modification. Among the most common molecules in this category are METTL3 and METTL14, both of which catalyze m6A methylation of mRNA (and other nuclear RNAs) both in vitro and in vivo. Additionally, WTAP serves as another key component within this methyltransferase complex.
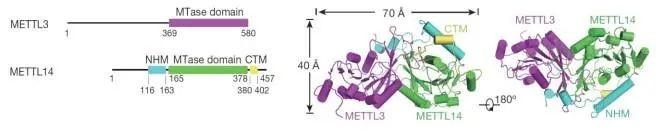
Structural biology studies have elucidated that METTL3 and METTL14 possess crucial catalytic domains, and they form a heterocomplex. METTL3 serves as the catalytic subunit, while METTL14 plays a pivotal role in substrate recognition. Additionally, WTAP, Vir, and other types of factors are integral components of this complex. Notably, WTAP plays a crucial role in recruiting METTL3 and METTL14. These proteins catalyze adenosine methylation both in vivo and in vitro. Homologous proteins resembling METTL3 and METTL14 have been discovered not only in mammals such as humans and mice but also in other organisms including fruit flies, yeast, and even Arabidopsis thaliana.
m6A Demethylases (Erasers)
Erasers function to "erase" RNA methylation modifications, thereby mediating the process of demethylation modification on RNA. In eukaryotes, the identified m6A demethylases primarily include FTO and ALKBH5. FTO and ALKBH5 are capable of removing m6A methylation from mRNA (and other nuclear RNAs).
 (Ying Yang et al (2015)
(Ying Yang et al (2015)
FTO protein shares structural similarities with the Alkb protein family in its core domain, yet it possesses a unique C-terminal long loop distinct from other Alkb family proteins. It is this distinctive domain that enables FTO protein to catalyze demethylation modifications on methylated single-stranded DNA or RNA. Dysregulation of FTO gene transcription levels can lead to various diseases such as acute myeloid leukemia. Another crucial demethylase, ALKBH5, is capable of demethylating mRNA in the nucleus. It harbors an N-terminal region rich in alanine and a unique coiled-coil structure. Knockdown of ALKBH5 in cell lines results in a significant increase in m6A modification levels on mRNA.
m6A Methylation Readers
Readers: These proteins "read" the information encoded by RNA methylation modifications and participate in downstream processes such as translation and degradation of RNA.
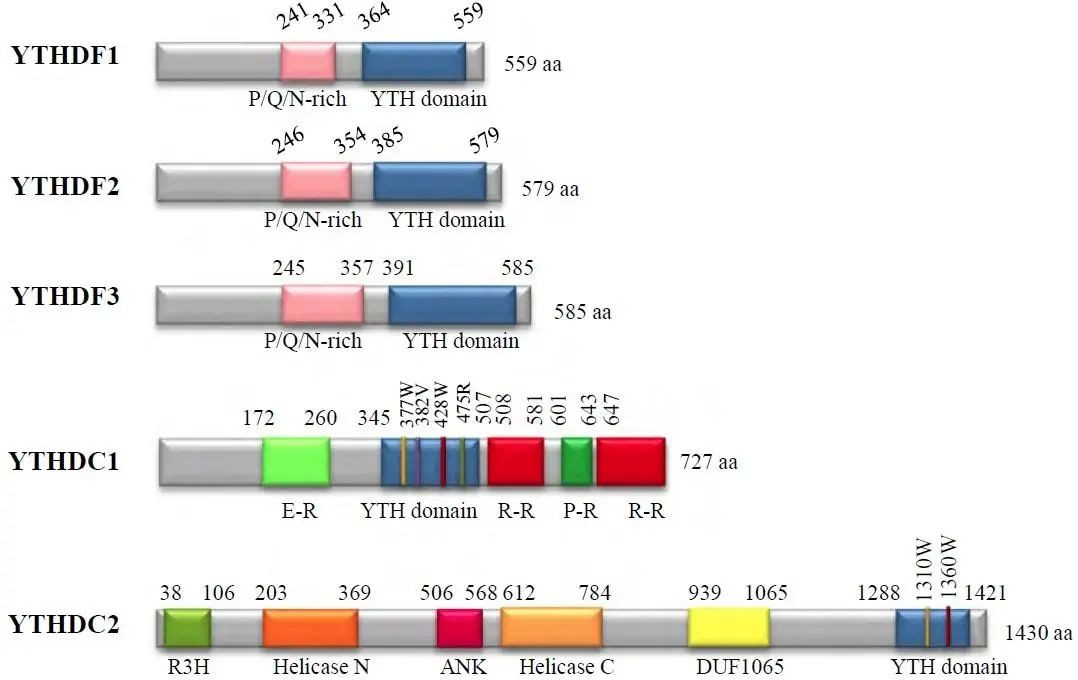
Various reader proteins have been identified through RNA pull-down experiments, including proteins containing YTH domains, heterogeneous nuclear ribonucleoproteins (hnRNPs), and eukaryotic initiation factors (eIFs). The functions of these reader proteins primarily involve specific binding to m6A-methylated regions, weakening homologous binding with RNA-binding proteins, and altering RNA secondary structure, thereby modulating protein-RNA interactions.
 (Ying Yang et al (2015)
(Ying Yang et al (2015)
Proteins containing the YTH domain include YTHDC1-2 and YTHDF1-3, among others. YTHDF1-3 primarily recognize m6A-modified mRNA in the cytoplasm, while the functions of YTHDC1-2 are predominantly localized to the nucleus. These proteins possess a YTH domain at the C-terminus, allowing for specific RNA binding through overlapping with the m6A motif. Additionally, their proline/glutamine/asparagine-rich (P/Q/N-rich) domains are associated with subcellular localization.
eIF3 proteins can bind to bases undergoing m6A modification in the RNA 5'UTR, thereby promoting mRNA translation. This represents a novel mechanism almost independent of traditional eIF4 activation for translation initiation. RNA recruited under the action of eIF3 forms a protein complex at the 5' end cap, facilitating the recruitment of the 43S ribosome.
HNRNPA2B1, a member of the hnRNP protein family, retains the functionality of a reader protein. Unlike YTHDC1 protein, HNRNPA2B1 does not directly bind to m6A-modified bases. Besides activating downstream pathways of primary miRNA (pri-miRNA), HNRNPA2B1 is also involved in pre-miRNA processing.
 (Ying Yang et al (2015)
(Ying Yang et al (2015)
How Do You Detect m6A?
Detection methods for m6A are diverse, including MeRIP-seq, miCLIP-seq, MeRIP-qPCR, LC-MS/MS, among others.
MeRIP-seq: Based on immunoprecipitation enrichment technology, this approach combines IP and input libraries for m6A modification analysis. The input library, akin to RNA-seq libraries, facilitates expression profiling analysis. Technical features: Simple library construction, mature technology.
miCLIP-seq: Utilizing UV cross-linking and IP technology, this method achieves single-base resolution detection. It accurately identifies m6A modification sites using CT mutations and can detect multiple m6A sites within a single peak. Technical features: Single-base resolution, requiring high RNA quality and input, complex library construction.
MeRIP-qPCR: Targeting specific regions for m6A modification detection, this method offers relatively high accuracy. It requires the addition of fully methylated and unmethylated standards. Technical features: Targeted approach, higher accuracy.
Research Strategy for m6A
Identification of Relevant Relationships:
Utilizing high-throughput sequencing technologies to comprehensively characterize the m6A methylation landscape:
Tracking changes in m6A peak abundance
Assessing alterations in m6A-modified gene numbers
Analyzing the distribution of m6A peaks within individual genes
Investigating the distribution of m6A peaks across genomic elements
Conducting motif analysis of m6A peaks
Performing functional analysis of genes modified by m6A peaks
Selection of Specific Differential m6A Peaks and Genes:
Identification of differential m6A peaks
Functional analysis of genes with differential m6A modifications
Protein-protein interaction (PPI) analysis of genes with differential m6A modifications
Analysis of m6A modifications in candidate genes
Integration of m6A Methylome and Transcriptome:
Correlation analysis between Differentially Methylated Genes (DMG) and Differentially Expressed Genes (DEG)
Selection of target genes modified by m6A
Verification of Causal Relationships:
Knockout or overexpression of m6A-related enzymes combined with transcriptome sequencing to identify target genes
Validation of gene function through knockout and overexpression of target genes
Confirmation of direct regulation of target genes by relevant methyltransferases through motif point mutation analysis
FAQ:
What are the effects of m6A modifications on mRNA?
m6A modifications exert diverse effects on mRNA. They can modulate mRNA stability by either promoting degradation or enhancing stability. Furthermore, m6A modifications play a role in regulating mRNA translation efficiency, influencing mRNA splicing patterns, mediating mRNA export from the nucleus to the cytoplasm, and controlling mRNA localization within cells.
For more information, refer to "What is the role of RNA m6A methylation?"
What diseases are associated with m6A?
Dysregulation of m6A modifications is implicated in various diseases. These encompass cancer, where altered m6A patterns contribute to tumor initiation, progression, and drug resistance. Neurological disorders like Alzheimer's disease and Parkinson's disease also exhibit associations with m6A dysregulation. Moreover, metabolic disorders such as obesity and diabetes, along with viral infections, are linked to disruptions in m6A-mediated processes.
References:
- Wang, S., Lv, W., Li, T. et al. Dynamic regulation and functions of mRNA m6A modification. Cancer Cell Int 22, 48 (2022).
- Wang, X., Feng, J., Xue, Y. et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 534, 575–578 (2016).
- Natalia Pinello, Stephanie Sun and Justin Jong-Leong Wong. Cancer Biology & Medicine November 2018, 15 (4) 323-334


 Sample Submission Guidelines
Sample Submission Guidelines
