Whole Exome vs. Whole Genome Sequencing
Next Generation Sequencing (NGS), also known as high-throughput sequencing, is characterized by high output and high resolution, and is capable of reading hundreds of thousands to millions of DNA molecules at a time in parallel. Therefore, it has become one of the most commonly used genetic tests with low cost, high accuracy, high throughput and rapid detection.
The utilization of genetic variants identified through both whole exome sequencing (WES) and whole genome sequencing (WGS) has seen a significant rise in both scientific research and clinical diagnosis.
What Is an Exon?
Exons represent crucial segments within eukaryotic genes, primarily tasked with encoding proteins. Throughout transcription, exons undergo RNA splicing, culminating in the formation of mature mRNA molecules. These mRNA molecules serve as templates for protein synthesis, thereby facilitating the fundamental process of gene expression. An exome encompasses the entirety of exonic regions within a genome, embodying the genetic blueprint for protein-coding elements.
What Is Exome Sequencing?
Whole exome sequencing (WES) involves enriching DNA specifically within exonic regions of the entire genome using sequence capture or targeting techniques, followed by high-throughput sequencing. In comparison to whole-genome sequencing, WES exhibits superior targeting precision, leading to reduced sequencing time and costs while achieving a deeper sequencing depth within genomic protein-coding regions. This results in more precise genotyping outcomes and enhances the detection of disease-associated rare mutations, making it particularly suitable for large-sample size studies on complex genetic diseases and cancer cohorts.
Presently, exome sequencing technology is extensively employed in identifying causative and susceptibility genes linked to diverse complex diseases, playing a crucial role in developing effective prevention and treatment strategies. Moreover, exome sequencing finds application in cancer research, studies involving normal populations, and investigations into hereditary diseases.
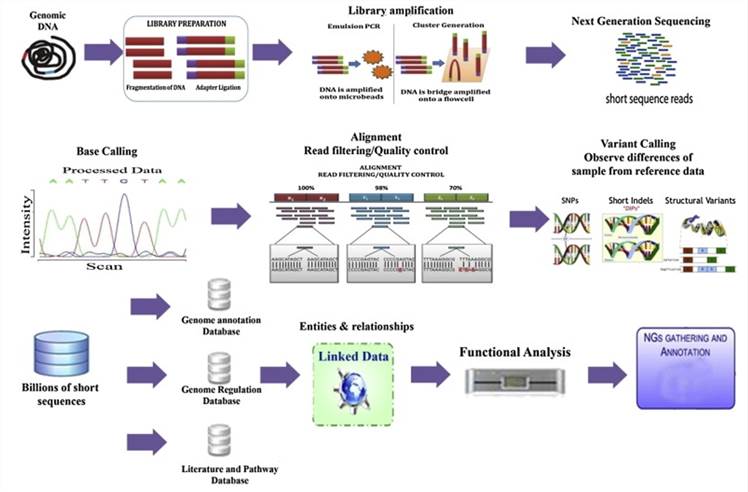 Next-Generation Sequencing methodology. (Nayarisseri et al., 2013)
Next-Generation Sequencing methodology. (Nayarisseri et al., 2013)
Recommended Reading: A Guide to Cancer Whole Exome Sequencing.
Advantages of Exome Sequencing
- Utilization of multiple annotation databases enhances the identification of deleterious mutations, complemented by various screening methods to minimize the risk of overlooking such mutations.
- Tailored protocols offer flexibility and precision aligned with specific research objectives.
- Rigorous adherence to stringent data quality control standards ensures the reliability and integrity of sourced data.
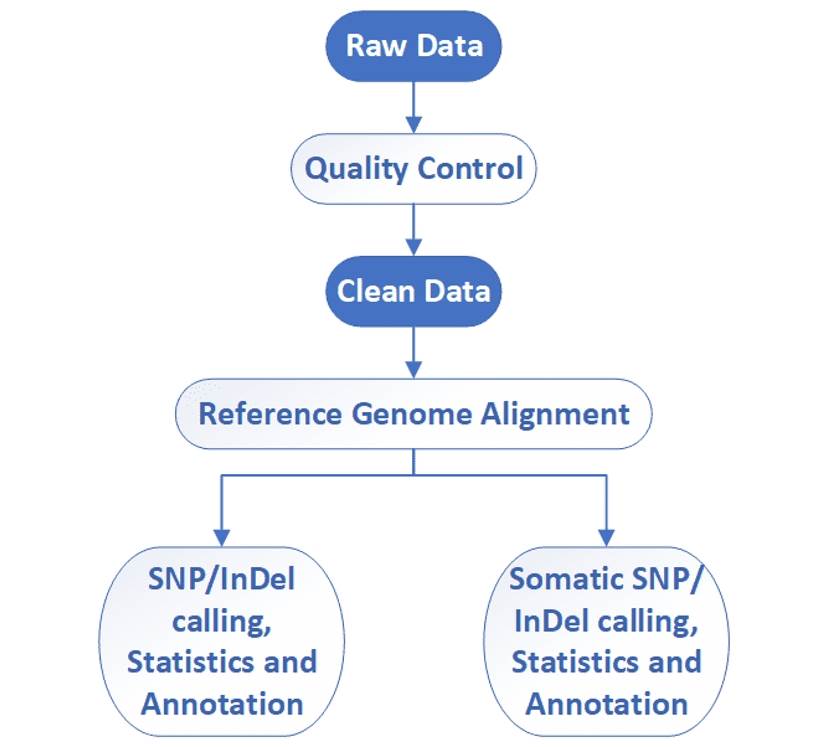 Whole exome sequencing - CD Genomics
Whole exome sequencing - CD Genomics
CD Genomics provides full whole exome sequencing service package including sample standardization, exome capture, library construction, deep sequencing, raw data quality control, and bioinformatics analysis.
CD Genomics high-throughput sequencing and long-read sequencing platforms facilitate the robust analysis of genome. This advanced sequencing approach allows for detecting common and rare single nucleotide polymorphisms (SNPs), copy number variations (CNVs) and other genetic variations, providing valuable insights into disease related genetics, individualized health management, breeding, etc.
What Is Genome Sequencing?
Whole human genome resequencing entails sequencing the entire genome of an individual or a population against the reference sequence of the human genome. This approach aims to screen for genetic variation across the entire genome, enabling analyses of genotypic diversity, genetic evolution, identification of disease-causing and susceptibility genes, single-gene disease screening, as well as cancer screening, among other applications.
Advancements and widespread adoption of next-generation sequencing technology have elevated whole-genome resequencing to a pivotal method in human genetics, translational medicine, and population evolution studies. This method enables comprehensive exploration of genome-wide sequence disparities and structural variations, encompassing single-base mutations, insertion-deletion variations, copy number variations, and structural variations (SNVs, SNPs, InDels, CNVs, and SVs). By scanning the entire genome, it facilitates the identification of mutation sites associated with phenotypic disparities, diseases, evolutionary processes, and more.
CD Genomics combines both Illumina HiSeq and PacBio systems to provide a fast and accurate whole genome sequencing and bioinformatics analysis for any species. Our highly experienced expert team executes quality management, following every procedure to ensure confident and unbiased results.
Advantages of Genome Sequencing
Whole Genome Sequencing (WGS) offers a multitude of advantages as a comprehensive method for analyzing the entire genome. Its primary strength lies in its ability to provide a thorough examination of genetic variation across all regions of the genome. Unlike targeted sequencing approaches, WGS encompasses both coding and non-coding regions, making it particularly valuable for studies focusing on non-coding variants. Recent research has underscored the significance of non-coding DNA variants in the manifestation of complex diseases, emphasizing the importance of WGS in uncovering these associations.
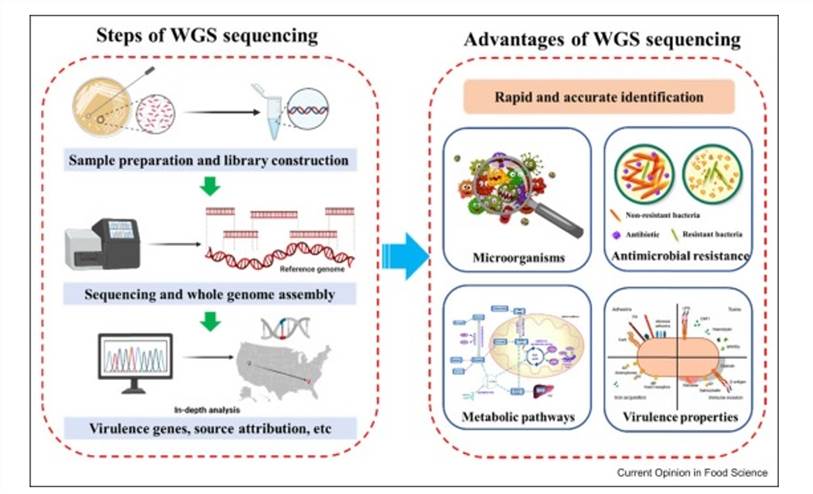 Whole-genome sequencing for food safety (Zhang et al., 2022)
Whole-genome sequencing for food safety (Zhang et al., 2022)
Moreover, WGS serves as the technology of choice for investigations targeting structural variants, offering insights into large-scale genomic alterations. With a typical sequencing depth exceeding 30X, WGS generates rich and detailed data, providing a comprehensive view of genomic variation within a sample. This depth of coverage enables the sensitive detection of genetic variants, including rare and low abundance mutations.
Recommended Reading: How to Decide Between 100X Whole Exome Sequencing (WES) and 30X Whole Genome Sequencing (WGS)?
Despite its advantages, WGS does present challenges, such as the substantial data volume generated, the associated high cost, and the complexity of data interpretation. However, as sequencing technologies continue to advance and costs decline, WGS is increasingly utilized in diverse fields beyond basic research. Its unparalleled capacity to uncover genetic variation and its implications for health and disease make WGS an indispensable tool for advancing genomic medicine and personalized healthcare.
Whole Exome Sequencing vs. Whole Genome Sequencing
Table 1 WES vs. WGS
| Whole Genome Sequencing | Whole Exome Sequencing | |
| Methodology | Sequences the entire genome | Selectively sequences exonic regions of the genome |
| Coverage | Comprehensive, including coding and non-coding regions | Focuses on protein-coding regions (exons) |
| Sequencing Depth | ≥ 30X | ≥ 100X |
| Advantages | - Genome-wide detection of mutation sites - Can detect all types of variants including CNVs/SVs - Does not require prior knowledge of the disease |
- Targets coding regions, facilitating mutation detection that alters protein products - Data analysis is less complex and easier to interpret - High sequencing depth enables sensitive detection of low abundance variants |
| Disadvantages | - Large amount of sequencing data, leading to higher costs - More challenging to interpret data comprehensively - Low sequencing depth may hinder detection of low abundance mutations required for specialized analyses such as fusion gene detection or viral integration analysis - Mutations identified may not always be actionable |
- Unable to detect variants in non-coding regions - Incomplete and potentially inaccurate detection of structural variants |
| Applications | - Broad exploration of genetic variation - Studies focusing on non-coding and structural variants - Investigation of complex diseases |
- Identification of known pathogenic mutations - Protein structure variation analysis - Common disease studies |
| Recommendation | - When in-depth genomic analysis is required - For studies with ample budget and computational resources - Where broader understanding of genetic variation is necessary |
- When focusing on known pathogenic mutations - For efficient and targeted genetic analysis - For cost-effective and high-throughput sequencing - Where coverage of exonic regions is sufficient for the study |
Choosing Between WES and WGS
If cost were not a limiting factor, whole genome sequencing (WGS) would undoubtedly emerge as the standard approach for detecting genomic variation. WGS offers the unparalleled advantage of comprehensively sequencing DNA, providing researchers with a complete picture of the genetic landscape within a sample. This capability is essential for exploring the impact of alterations in non-coding regions on cellular function and for identifying large chromosomal variations, including structural and copy number variations.
Despite its capabilities, WGS comes with a significant drawback: its high cost. Additionally, a challenge lies in the vast number of variants detected, many of which may not be relevant to the disease or research question at hand. Thus, meticulous filtering is necessary to sift through these variants and discern the ones of interest. However, as sequencing costs continue to decline over time, the utility of WGS extends beyond basic research, finding applications in various fields.
The decision between WES and WGS depends on the specific research or clinical objectives, budget constraints, and computational resources available. For a more detailed comparison, please see our Table 1 or you can refer to our article WGS vs. WES vs. Targeted Sequencing Panels.
- WGS offers a comprehensive view of the genome and is suitable for studies requiring broad exploration of genetic variation or discovery of novel variants.
- WES, with its focused approach and lower cost, is preferred for targeted analyses aiming to identify disease-causing mutations, especially in known coding regions.
Choosing between WGS and WES depends on the specific research objectives, budget constraints, and the desired depth of genomic analysis. WGS provides a comprehensive view of the genome, making it suitable for studies requiring broad exploration of genetic variation. On the other hand, WES offers a targeted approach, focusing on protein-coding regions, and is preferred for identifying known pathogenic mutations. Researchers should carefully consider the advantages and limitations of each method to select the most appropriate approach for their study.
References:
- Nayarisseri, Anuraj, et al. "Impact of Next-Generation Whole-Exome sequencing in molecular diagnostics." Drug Invention Today 5.4 (2013): 327-334.
- Zhang, Runrun, et al. "Whole-genome sequencing: a perspective on sensing bacterial risk for food safety." Current Opinion in Food Science 47 (2022): 100888.


 Sample Submission Guidelines
Sample Submission Guidelines
