Transposable Elements (TEs) in Eukaryotes
Transposable Elements (TEs) are virtually ubiquitous across species and exert a profound influence on genome structure and species evolution. However, it's noteworthy that TEs have historically received relatively limited attention. In the initial 60 years following their discovery, only a few TEs were extensively characterized, primarily within model organisms and select agricultural crops. These studies mainly delved into the phenotypic effects of TEs.
In the past decade, advances in genomic sequencing technologies have afforded us the ability to comprehensively identify, compare, and delve into the regulatory and transcriptional activities of various TEs. This has enabled a more intricate exploration of TEs at a finer granularity. A notable revelation has been that the majority of TEs within genomes are inactivated transposon remnants, akin to dormant volcanoes, with the cumulative presence of these relics constituting the repetitive sequences found in eukaryotic genes.
What Are Transposable Elements?
Transposable elements, commonly referred to as TEs, are specialized DNA sequences that possess the remarkable ability to autonomously relocate within the host genome. These sequences typically span a range of 100 to 10,000 base pairs in length. Much like viruses, TEs exhibit a complex and self-serving nature, often encoding proteins with multifaceted biochemical functions or housing non-coding regulatory segments that play a crucial role in the transposition process.
In the context of eukaryotes, TEs are prevalent in the genomes of nearly all eukaryotic species, with only a few exceptions. In certain species, the TE content can account for a substantial portion of the genome, potentially reaching up to 85%. Notably, the extent of TEs in a species remains unrelated to the complexity of its genome. Complex multicellular organisms, such as conifers and salamanders, can harbor substantial quantities of TEs, but intriguingly, certain unicellular organisms like Trichomonas vaginalis and Ancaliia algerae also exhibit elevated TE levels.
Categorizing Eukaryotic TEs
In 1989, David Finnegan introduced an initial classification scheme for TEs, dividing them into two primary classes:
- Class I: Retrotransposons
- Class II: DNA transposons
Class I retrotransposons employ RNA as an intermediary, facilitating the reverse transcription of RNA into DNA. This process culminates in the integration of the DNA copy into the host genome, often likened to a "copy-and-paste" mechanism. In contrast, Class II transposons primarily employ a "cut-and-paste" mechanism, accomplishing transposition without the involvement of an RNA intermediary.
Further refinement of TE classification is possible, taking into account the specific mechanisms of TE replication and integration. These two primary classes can be further subdivided into numerous subclasses, and upon deeper examination, they can be categorized into superfamilies, families, and so forth.
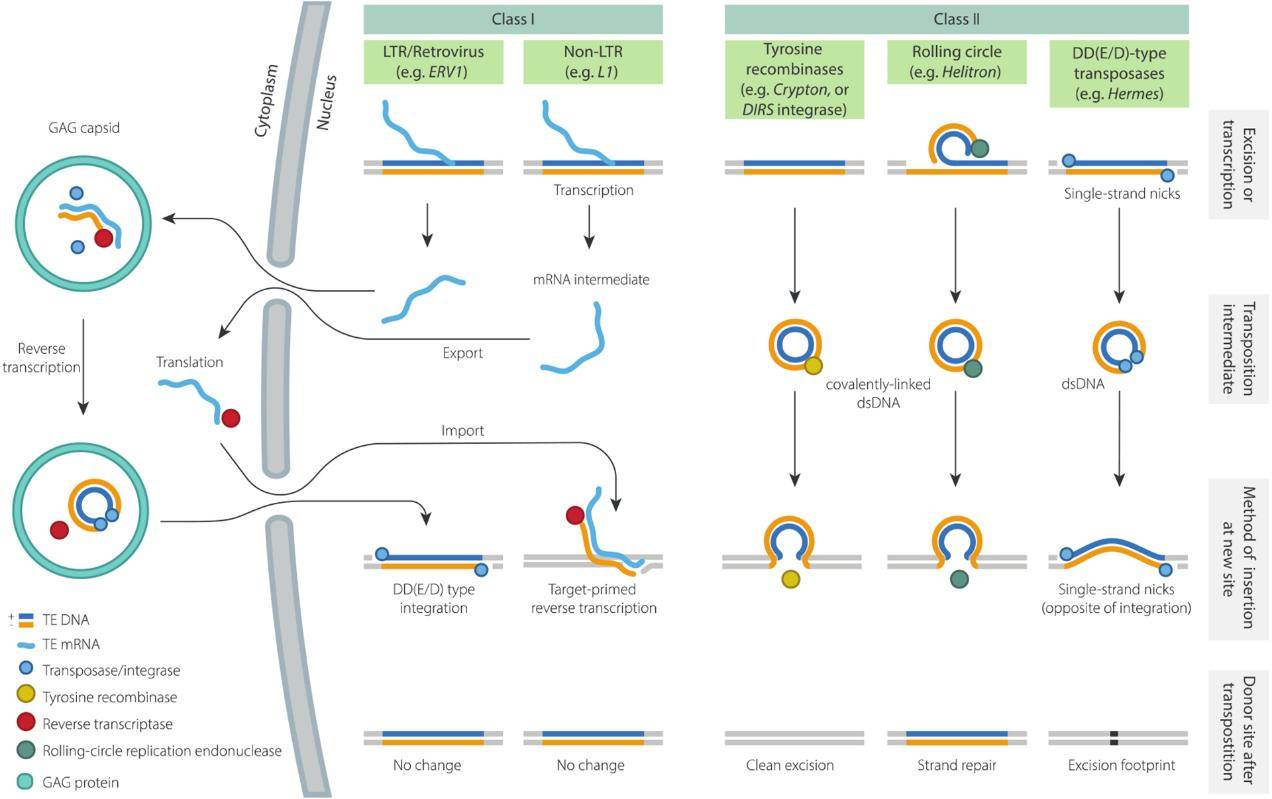 Summary of replication mechanisms and transposition intermediates. (Wells et al., 2020)
Summary of replication mechanisms and transposition intermediates. (Wells et al., 2020)
Disparities in Transposon Distribution Across Species
The prevalence of transposable elements (TEs) in genomes displays significant variability among different species. Some species have a scarcity of TEs in their genomes, while others are abundantly populated by these genetic elements.
Several studies have proposed a correlation between TE abundance and the effective population size of a species. A larger effective population size tends to heighten the efficiency of natural selection, thereby intensifying selective pressure on TEs. For instance, in species like Drosophila, which boast a sizable effective population size, TE content remains relatively low. In contrast, vertebrates, characterized by smaller effective population sizes, exhibit a reduced selective pressure on TE insertions, which can readily proliferate within the population. However, it's worth noting that even among species with similar effective population sizes, TE content can differ significantly. Therefore, differences in effective population size alone do not suffice to elucidate the variations in TE distribution.
Furthermore, TE distribution diverges markedly based on the type of TE. For instance, long terminal repeat elements (LTRs) are prevalent in flowering plants, non-LTRs are widespread in mammals, and DNA transposons are extensively distributed in organisms like zebrafish and nematodes. Hence, disparities in the prevalence of these elements cannot be solely attributed to variations in effective population sizes.
In summary, the intricate dynamics of TE distribution result from a combination of factors that extend beyond the size of the effective population, encompassing intricate genetic and ecological interactions specific to each species.
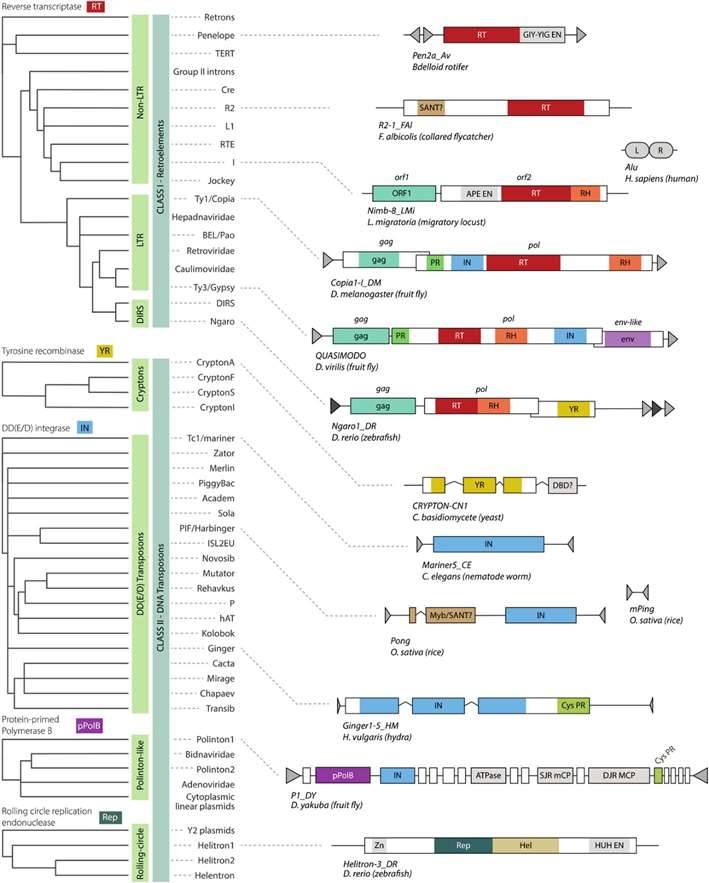 Structure and taxonomy of eukaryotic TEs. (Wells et al., 2020)
Structure and taxonomy of eukaryotic TEs. (Wells et al., 2020)
Transposon Abundance and Genome Size Nexus
Transposable elements (TEs) are a ubiquitous presence in the genomes of nearly all eukaryotic species, with only a handful of exceptions, primarily among unicellular organisms such as Plasmodium, Toxoplasma gondii, Microsporidium enteroencephalitis. Notably, the exceptions are characterized by exceptionally compact genomes, hinting at a potential correlation between genome size and TE abundance. However, it is essential to recognize that this relationship is more intricate than a mere direct proportionality.
To illustrate this complexity, consider the case of Anncaliia algerae, a unicellular organism with a modest genome size of 23 Mb, of which a substantial 14% comprises TEs, encompassing 240 TE families.
In larger genomes, such as that of salamanders, which can reach sizes of up to 120 Gb, the enlargement is primarily driven by the incorporation of LTR transposable elements. Plant genomes similarly undergo significant expansion due to transposable elements. While these elements may span a wide array of TE families, individual TEs can play an extraordinarily pivotal role. For instance, the brown hydroid, diverging from the green hydroid approximately 36 million years ago, experienced a rapid genome size increase from 300 Mb to 1 Gb, largely attributed to the CR1 non-LTR transposon.
Another factor influencing genome size and TE abundance is the deletion of nonessential DNA. Salamanders, for instance, exhibit larger genomes partly due to lower DNA deletion rates. In contrast, in species like Arabidopsis and rice, high rates of genome deletion, driven by ectopic recombination, counteract the genome enlargement resulting from transposition. This intricate balance sustains genome size stability in Arabidopsis and rice, and analogous phenomena are observed in birds and mammals.
In essence, the interplay between genome size and TE abundance is multifaceted, influenced not only by the presence of TEs but also by the processes of TE-driven expansion and DNA deletion, all of which shape the genomes of various species in unique ways.
Transposable Element Diversity
Transposable elements (TEs) exhibit a profound diversity in their distribution among species, extending beyond mere differences in abundance. The complex interplay between hosts and TEs often results in the formation of intricate TE family structures, giving rise to expanded TE subfamily species, as exemplified by L1 elements. Additionally, certain transposable elements, such as Helitrons, can generate new subfamilies by incorporating fragments of host DNA into their makeup.
TE distribution in eukaryotes is remarkably diverse, regardless of the measurement scale. For instance, zebrafish, known for its unparalleled TE abundance and diversity among vertebrate model organisms, harbors nearly 2,000 TE families spanning all subclades and encompassing nearly all superfamilies. Notably, zebrafish boasts a particularly high prevalence of DNA transposons, with approximately 1,000 distinct DNA transposon families that originated at different points in time, an unusual characteristic within the realm of fish genomes.
However, it's crucial to recognize that larger genome sizes do not inherently translate to greater TE diversity. A compelling illustration of this can be found in spruce, a gymnosperm with a genome size of 20 Gb. While it indeed harbors a substantial number of TE copies, these are predominantly concentrated within the LTR superfamily, with the majority of these transposons emerging between 5 and 60 million years ago. In contrast, species like rice and maize predominantly feature transposons younger than 5 million years. This contrast suggests that, in the case of spruce, even though TE diversity may appear relatively low, many of the pre-existing TEs in the genome have gradually diminished over time. Conversely, in many flowering plants, TE diversity remains high despite their comparatively small genomes. Intriguingly, there is a noticeable negative correlation between genome size and TE diversity in terrestrial plants.
In essence, the intricate mosaic of TE diversity reflects a dynamic interplay between genetic elements and host genomes, resulting in a tapestry of genomic diversity across the eukaryotic spectrum.
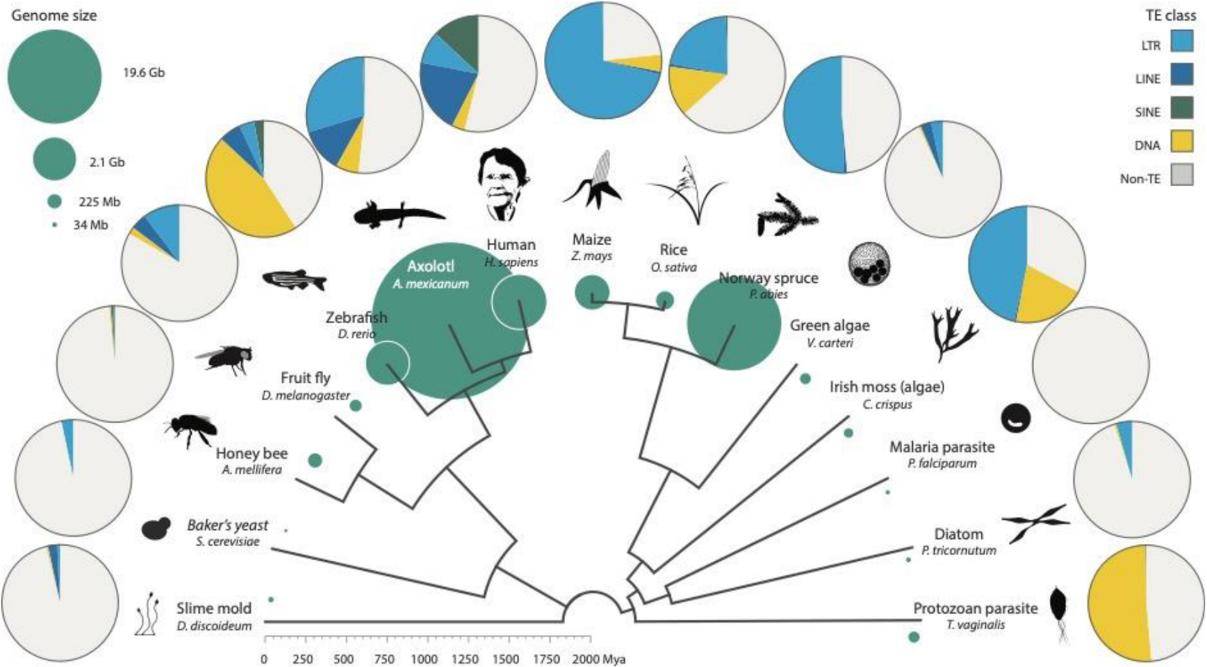 Distribution of TEs across the eukaryote phylogeny. (Wells et al., 2020)
Distribution of TEs across the eukaryote phylogeny. (Wells et al., 2020)
An in-depth study of TEs should inherently encompass an examination of their transposition mechanisms. However, it's increasingly common to observe investigations at the genomic evolution level while overlooking the fundamental transposition mechanisms of TEs themselves.
TE families exhibit substantial variability among themselves, and their impact on host genomes is of significant consequence. Therefore, an imperative focus is to identify and categorize TEs across a broader spectrum of species. Simultaneously, there's a growing need for concentrated research on the transposition mechanisms, particularly those of more specialized transposons such as Helitrons, Mavericks, YR elements, and the like.
In summary, the realm of Transposable Elements is a pivotal domain of study that offers valuable insights into genetic evolution. Comprehensive exploration, spanning both the diversity of TE families and the underlying transposition mechanisms, is crucial for a holistic understanding of their role in shaping genomes and driving species evolution.
Reference:
- Wells, Jonathan N., and Cédric Feschotte. "A field guide to eukaryotic transposable elements." Annual review of genetics 54 (2020): 539-561.


 Sample Submission Guidelines
Sample Submission Guidelines
