Single-Cell Transcriptome Research Methods and Applications
The field of genomics has undergone a revolutionary transformation with the advent of single-cell transcriptomics. Traditional bulk RNA sequencing techniques provide valuable insights into the average gene expression levels of a population of cells, masking the heterogeneity that exists within. Single-cell transcriptomics has emerged as a powerful tool for dissecting this heterogeneity by examining the transcriptome of individual cells. This breakthrough technology allows scientists to delve into the intricacies of gene expression, providing a comprehensive view of cellular diversity and function at an unprecedented level of resolution.
Single-cell transcriptomics, often referred to as single-cell RNA sequencing (scRNA-seq), is a methodology that enables the profiling of the transcriptome in individual cells. The technique was born from the merger of two critical dvancements: advances in single-cell isolation and sequencing technology.
In the early 2000s, the development of microfluidic devices and droplet-based systems allowed for the isolation of single cells from a heterogeneous population efficiently. Single cells are captured, lysed, and their RNA content preserved in these devices. The captured RNA is then reverse-transcribed into complementary DNA (cDNA) and amplified for sequencing.
The second crucial element is the utilization of high-throughput sequencing technology, which has witnessed substantial advancements in terms of affordability and efficiency. Key sequencing platforms like Illumina and 10× Genomics have played a pivotal role in propelling the progression of scRNA-seq. These cutting-edge technologies possess the capacity to generate massive volumes of data from individual cells, thereby facilitating comprehensive investigations into gene expression patterns, isoform diversity, and even alternative splicing.
Single-Cell Transcriptome Methods:
Single-cell transcriptome research involves the study of gene expression at the single-cell level, which necessitates highly sensitive and precise techniques. There are two primary approaches to single-cell transcriptomics: single-cell RNA sequencing (scRNA-seq) and single-cell RNA fluorescence in situ hybridization (FISH).
1) Single-Cell RNA Sequencing (scRNA-seq):
a. Droplet-based scRNA-seq: This method employs microfluidic technologies to encapsulate single cells in nanoliter-sized droplets, where cell lysis, reverse transcription, and amplification of cDNA take place. Popular platforms like the 10× Genomics Chromium system have enabled high-throughput, cost-effective profiling of thousands of cells simultaneously.
b. Plate-based scRNA-seq: Unlike droplet-based methods, plate-based scRNA-seq allows for more flexibility in experimental design. Researchers can manually isolate and process individual cells in microwell plates, leading to higher capture efficiency and reduced costs for smaller-scale projects.
c. Smart-seq2: This protocol focuses on full-length cDNA amplification, offering superior sensitivity and the ability to detect low-abundance transcripts. It is particularly useful for studying rare cell types or when comprehensive gene coverage is essential.
2) Single-Cell RNA Fluorescence in Situ Hybridization (FISH):
FISH is a microscopy-based technique that directly visualizes individual RNA molecules in situ within intact cells, allowing for precise spatial and temporal information on gene expression. Various FISH-based methods, such as smFISH (single- molecule FISH) and MERFISH (multiplexed error-robust FISH), have been developed to investigate single-cell gene expression patterns.
Applications of Single-Cell Transcriptome
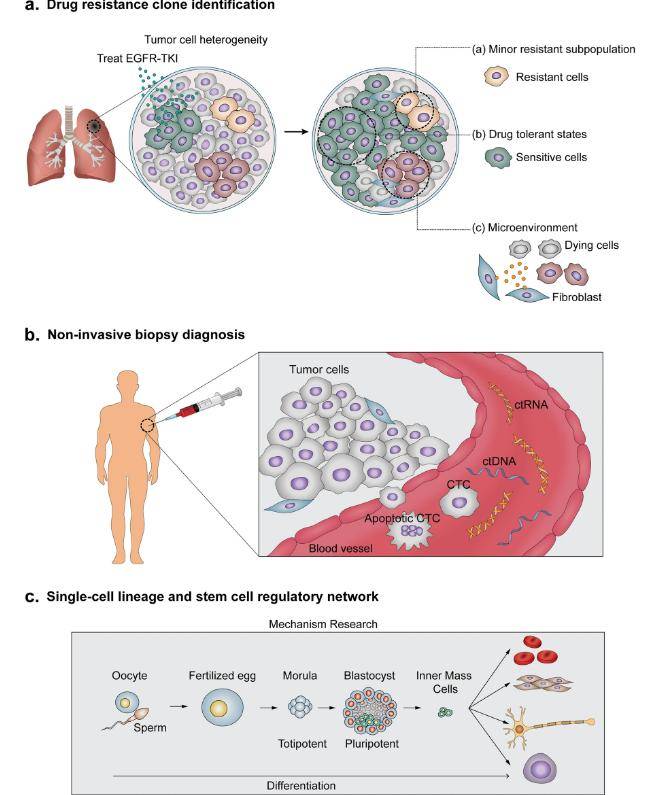 (Byungjin Hwang et al,. nature 2018)
(Byungjin Hwang et al,. nature 2018)
Exploring the transcriptomes at the single-cell level holds diverse applications spanning various biological disciplines. This endeavor is reshaping our comprehension of cellular biology, elucidating disease mechanisms, and advancing the field of therapeutic development.
a. Cellular Heterogeneity and Development:
Studying single-cell gene expression profiles has revealed the remarkable cellular diversity within tissues, including the nervous system, immune system, and embryonic development. Researchers can trace lineage progression, identify cell subtypes, and understand the role of specific genes in cellular differentiation.
b. Disease Mechanisms:
Single-cell transcriptomics is instrumental in uncovering the molecular mechanisms of diseases, such as cancer, neurodegenerative disorders, and autoimmune conditions. By comparing diseased and healthy tissues at the single-cell level, researchers can pinpoint the origins of pathology and potential therapeutic targets.
c. Immunology:
In immunology, single-cell transcriptome research has been pivotal in characterizing immune cell populations, their activation states, and immune responses. This knowledge is vital for the development of novel vaccines immunotherapies, and understanding autoimmune diseases.
d. Neuroscience:
The heterogeneity of neuronal cell types and their intricate connections in the brain have been elucidated using single-cell transcriptomics. This technique aids in identifying novel drug targets for neurological disorders and understanding how neural circuits function.
e. Stem Cell Biology:
Single-cell transcriptomics has advanced our comprehension of stem cell dynamics and their differentiation into various cell types. This knowledge is crucial for regenerative medicine and tissue engineering applications.
f. Single-Cell Diagnostics:
The potential for single-cell transcriptomics in personalized medicine is immense. Identifying specific gene expression patterns in individual patients can guide treatment decisions and predict therapy responses.
g. Drug Discovery:
Pharmaceutical companies are progressively utilizing state-of-the-art single-cell transcriptomics to explore potential drug targets and screen compounds with therapeutic efficacy. Concurrently, the real-time monitoring of drug responses at the single-cell level enables a more precise evaluation of drug efficacy and safety. Consequently, this approach facilitates the development of more effective treatment regimens for patients, concurrently minimizing adverse drug reactions. Undoubtedly, this innovative methodology holds the promise of revolutionary advancements in the pharmaceutical field.
h. Challenges and Future Directions:
While single-cell transcriptome research has made significant strides, it is not without challenges. Some key issues include data analysis complexity, high costs, and the need for standardized protocols. Researchers are continually working to address these challenges and further advance the field.
Challenges and Limitations
While single-cell transcriptomics has undeniably revolutionized genomics, it comes with its set of challenges and limitations:
Data Complexity: The high-dimensional data generated from scRNA-seq is complex and requires advanced computational tools for analysis. Bioinformatics expertise is crucial for extracting meaningful insights.
Cost and Throughput: Single-cell sequencing can be expensive, and the number of cells that can be sequenced in a single experiment is limited, making large-scale studies costly and time-consuming.
Technical Variability: There can be variability introduced at various stages of the scRNA-seq workflow, from cell capture to library preparation. Quality control measures are necessary to minimize such technical artifacts.
Data Integration: Integrating data from different experiments or platforms can be challenging due to technical and biological variations. Methods for harmonizing datasets are actively evolving.
Future directions in single-cell transcriptomics involve the integration of multi- omics data (e.g., genomics, epigenomics, proteomics) to provide a comprehensive view of cellular function. Additionally, spatial transcriptomics techniques are emerging, allowing for the mapping of gene expression within tissue sections, which can provide critical insights into cell-cell interactions and microenvironmental influences.
Single-cell transcriptome research has revolutionized our understanding of cellular biology, development, disease, and a wide range of biological processes. Through techniques like scRNA-seq and single-cell FISH, researchers can explore the heterogeneity within tissues, leading to advancements in diagnostics, therapeutics, and our overall knowledge of biology. As technology and analytical methods continue to improve, single-cell transcriptomics will remain a pivotal tool in the arsenal of modern biology, promising even more groundbreaking discoveries in the years to come.
References:
- Stuart, T., & Satija, R. (2019). Integrative single-cell analysis. Nature Reviews Genetics, 20(5), 257-272.
- Shalek, A. K., & Benson, M. (2017). Single-cell analysis: Progress, challenges, and opportunities. Genome Biology, 18, 84.
- Cao, J., & Ramani, V. (2021). Understanding cellular heterogeneity with single-cell RNA sequencing. Nature Reviews Genetics, 22(9), 479-492.
- Xia,Y., & Bushman, F. D. (2019). Obstacles and opportunities in applying single-cell methods to HIV reservoirs. Trends in Microbiology, 27(8), 684-696.
- Potter, S. S. (2018). Single-cell RNA sequencing for the study of development, physiology, and disease. Science, 360(6385), 1457- 1462.


 Sample Submission Guidelines
Sample Submission Guidelines
