Microarray vs. RNA Sequencing
The transcriptome encompasses the entirety of RNA species within a cell, comprising mRNAs, rRNAs, tRNAs, both degraded and intact forms. Dissecting this transcriptomic landscape is crucial for unraveling the intricacies of cellular function. Among the technologies devised for this purpose, microarrays and RNA sequencing stand out prominently.
RNA sequencing and microarrays are prominent tools for genome-wide transcriptome analysis, each with its distinct advantages and mechanisms. A fundamental divergence lies in their operational principles: microarrays rely on the hybridization affinity of predefined labeled probes to the target cDNA sequence, whereas RNA sequencing employs direct sequencing of the cDNA strand through cutting-edge technologies like NGS.
Microarrays necessitate prior knowledge of the sequence, whereas RNA sequencing operates independently of such foreknowledge. Below, we delve into the nuanced disparities between these two methodologies.
CD Genomics high-throughput sequencing and microarray services enable in-depth analysis of transcriptomes.
What is Gene Expression Microarray?
Microarray technology has revolutionized the study of gene expression dynamics, offering unparalleled insights into the intricate mechanisms governing cellular processes. Among its diverse applications, RNA microarray stands out as a pivotal method for probing RNA expression levels with precision and efficiency. Here, we delve into the workings of the microarray, shedding light on its methodology and transformative potential.
At its core, microarray harnesses the versatility of microarray chips to interrogate the transcriptional landscape of genes. The process commences by immobilizing known gene templates onto a vector, establishing a foundation for subsequent analysis. This foundational step lays the groundwork for exploring a myriad of objectives, including unraveling transcriptional regulation, identifying novel transcripts, and comparing differentially expressed genes across various samples.
Workflow of RNA Microarray
The essence of RNA microarray lies in its meticulous methodology, which orchestrates a symphony of molecular interactions to unveil gene expression profiles. Upon immobilization of gene templates, RNA is extracted from both treated and control samples, paving the way for reverse transcription into complementary DNA (cDNA). Employing distinct fluorescent dyes – red for Test and green for Reference – facilitates discrimination between samples, enabling comparative analysis with unparalleled precision.
The ensuing step in the RNA microarray process entails hybridizing the cDNAs to their respective gene templates, followed by meticulous elution of unbound cDNAs. This intricate dance of molecular interactions culminates in the examination of each locus under a laser, where the abundance of mRNA at each site is meticulously quantified. The resultant light signals, meticulously captured under laser illumination, serve as a testament to the dynamic interplay of gene expression within cellular ecosystems.
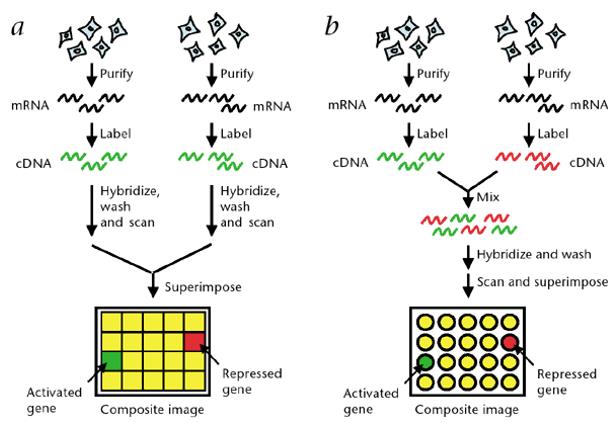 Expression analysis by microarray. (Stears et al., 2003)
Expression analysis by microarray. (Stears et al., 2003)
In the final analysis, RNA microarray unveils a tapestry of gene expression dynamics, offering invaluable insights into the molecular underpinnings of biological phenomena. The interpretation of these intricate signals is facilitated by the discerning eye of the observer, who discerns subtle variations in gene expression patterns. For instance, the appearance of gene A as a vibrant yellow hue signifies balanced expression across treatment and control groups, while the emergence of gene B in a deeper shade of red heralds a pronounced upregulation in the treatment group, exemplifying the nuanced insights afforded by microarray technology.
- Isolation of Total RNA: Commence by extracting total RNA from cells or tissues, laying the foundation for downstream analysis.
- Conversion of RNA to cDNA: Employ reverse transcriptase to transmute RNA into complementary DNA (cDNA), while concurrently tagging the cDNA molecules with fluorescent markers, facilitating subsequent detection.
- Hybridization onto Microarray: Engage in the pivotal step of hybridization, wherein the labeled cDNA molecules interface with a meticulously designed microarray chip. Each probe on the chip corresponds to a distinct gene or transcript, enabling comprehensive interrogation of gene expression dynamics.
- Scanning and Signal Acquisition: Employ scanning technology to capture the fluorescent signal intensity emanating from each probe on the microarray chip. This signal intensity serves as a direct reflection of the expression level of the corresponding gene or transcript within the sample under scrutiny.
- Data Analysis and Interpretation: data analysis utilizing specialized software, encompasses a spectrum of tasks including data preprocessing (e.g., normalization, background correction, etc.), quality assessment, and the pivotal task of differential expression analysis. Through meticulous data interpretation, uncover the nuanced intricacies of gene expression dynamics, propelling biological inquiry to unprecedented heights.
What is RNA-Seq?
RNA-Seq stands as the pinnacle of gene expression analysis, offering unparalleled depth and adaptability in probing genome-wide transcriptional dynamics. This groundbreaking technology is transforming our understanding of cellular processes with its precision and versatility.
At its core, RNA-seq, or RNA sequencing, is a cutting-edge method devised to decipher the sequence of RNA molecules within a biological sample. This revolutionary technique employs high-throughput sequencing of complementary DNA (cDNA) molecules synthesized from the target RNA pool through reverse transcription. Utilizing next-generation sequencing platforms, RNA-seq unveils the intricate sequence of cDNA, enabling precise elucidation of RNA sequences and their relative abundance across the genome.
CD Genomics high-throughput sequencing and library construction services enable in-depth analysis of transcriptomes.
The true power of RNA-seq lies in its unparalleled sensitivity and reliability. Capable of detecting even the most elusive and low-abundance RNA species, RNA sequencing empowers researchers to unravel the intricate tapestry of gene expression with unprecedented granularity. Moreover, its remarkable sensitivity renders it adept at identifying rare transcripts, shedding light on elusive biological phenomena previously obscured by conventional techniques.
Yet, perhaps the most remarkable aspect of RNA sequencing is its capacity to unveil novel RNA sequences and unravel the complexities of alternative splicing. Unlike traditional methods constrained by pre-existing knowledge, RNA-seq transcends boundaries, offering a comprehensive snapshot of the transcriptomic landscape, including previously undiscovered transcripts and splice variants.
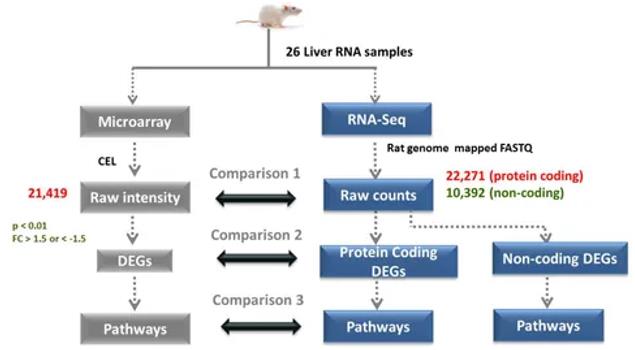 Overall computational process of RNA-Seq and microarray data analysis. (Rao et al., 2019)
Overall computational process of RNA-Seq and microarray data analysis. (Rao et al., 2019)
RNA-Seq vs. Microarray Technologies
RNA-seq and transcriptomics microarray technologies stand as pillars in the realm of gene expression analysis, each offering unique advantages and insights. Here, we delve into their similarities and divergences to elucidate their roles in deciphering the intricacies of the transcriptome.
Similarities
- Gene Expression Analysis: Both RNA-seq and microarrays serve as indispensable tools for probing gene expression patterns within biological samples.
- RNA Detection: They both enable the identification and quantification of RNA molecules present in a sample at a specific moment.
- Sequence Dependency: Both techniques rely on the sequence of RNA molecules for detection and analysis.
- Quantification: Both methods facilitate the determination of primary sequence and relative abundance of RNA species within a sample.
- Reproducibility: Both RNA-Seq and microarray technologies boast high levels of reproducibility, ensuring consistency and reliability in experimental results.
Advantages of RNA-Seq Over Microarray Technology
- Preference-Free Transcript Detection: RNA sequencing liberates researchers from the constraints of species- or transcript-specific probes, enabling the detection of novel transcripts, gene fusions, single nucleotide variants, and other previously elusive changes.
- Expanded Dynamic Range: RNA sequencing surpasses microarray technology in dynamic range, offering discrete quantification and digitized reads with a wider range (>105 for RNA-Seq vs. 103 for microarrays), thus overcoming limitations imposed by background noise and signal saturation.
- Enhanced Specificity and Sensitivity: RNA-seq exhibits superior specificity and sensitivity compared to microarrays, resulting in enhanced detection of genes, transcripts, and differential expression.
- Detection of Rare Transcripts: RNA-seq facilitates the detection of rare and low abundance transcripts with ease, allowing for increased sequencing coverage to unveil transcripts present at single-cell levels or weakly expressed genes.
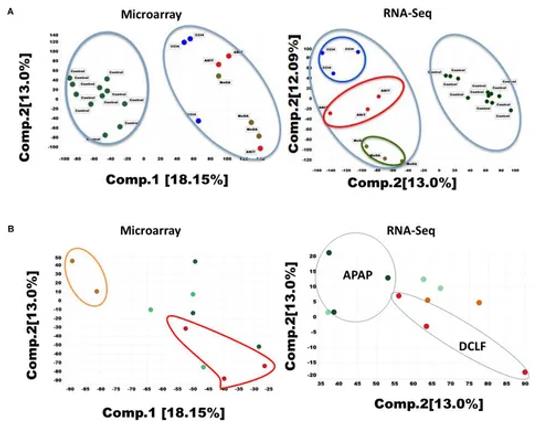 Principal component analysis (PCA) of the RNA-seq and microarray dataset for 26 liver samples. (Rao et al., 2019)
Principal component analysis (PCA) of the RNA-seq and microarray dataset for 26 liver samples. (Rao et al., 2019)
Summary
RNA sequencing, a sequencing-based approach, and microarrays, utilizing probe hybridization, serve as indispensable tools in transcriptome analysis. While microarrays excel in detecting known sequences, they falter in detecting novel or less common RNA sequences. In contrast, RNA-seq transcends these limitations, offering comprehensive coverage of all RNA sequences present in a sample. The key disparity between RNA-seq and microarrays lies in their detection methods.
RNA-seq's wider dynamic range and heightened sensitivity make it the preferred choice for discerning novel transcripts and unraveling intricate gene expression patterns. Nevertheless, the complementary nature of RNA-seq and transcriptomics microarray technologies allows for a comprehensive and nuanced exploration of the transcriptome, underscoring their collective importance in advancing our understanding of biological processes.
References:
- Rao, Mohan S., et al. "Comparison of RNA-Seq and microarray gene expression platforms for the toxicogenomic evaluation of liver from short-term rat toxicity studies." Frontiers in genetics 9 (2019): 425202.
- Stears, Robin L., Todd Martinsky, and Mark Schena. "Trends in microarray analysis." Nature medicine 9.1 (2003): 140-145.


 Sample Submission Guidelines
Sample Submission Guidelines
