How to Detect Transcription Factor Binding Sites (TFBS) by ChIP-Seq
What Are Transcription Factors?
Transcription factors are pivotal regulators of gene expression, orchestrating the intricate dance of genetic information within cells. With approximately 1,900 known transcription factors in the human genome, these molecular maestros play a critical role in determining cellular function and the response to environmental stimuli.
Transcription factors come in various flavors, each tailored to fulfill specific functions. While they possess DNA-binding domains as their hallmark feature, some also sport structural domains for dimerization or interactions with co-factors. Their distribution across tissues, activity levels, and regulation vary, making them essential cogs in the gene expression machinery. Broadly, transcription factors can be categorized into two groups: site-specific transcription factors and general transcription factors.
a. Site-Specific Transcription Factors
This category predominantly governs cell-specific gene expression, shaping the unique identities of different cell types.
b. General Transcription Factors
Comprising components like TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, these factors are essential components of the transcription machinery.
What is Transcription Factor Binding Site (TFBS)?
In a vast genome of approximately 3.26 billion base pairs, identifying the needle in the haystack becomes paramount. Here's where the concept of transcription factor binding sites (TFBS) comes into play. These sites are regions within the genome where transcription factors specifically interact to modulate gene expression.
Predicting TFBS with precision has been an enduring challenge. Computer simulations have, in some cases, overestimated binding sites by up to 1,000-fold. Recent advancements, fueled by the availability of vast experimental data, have significantly improved our ability to predict functional TFBS. Techniques such as ChIP-seq, which locates protein-DNA interactions in a genome-wide manner, have revolutionized our understanding of TFBS.
ChIP-seq: The Game Changer
Chromatin immunoprecipitation combined with sequencing (ChIP-seq) has emerged as a game-changing technique. It allows researchers to map protein-DNA interactions with unprecedented accuracy. By comparing this data with the reference genome, scientists can unravel genome-wide associations between specific transcription factors or histone modifications and DNA sequences.
A concrete example illustrates the power of ChIP-seq in understanding the regulation of transcription factors and their binding sites. In this study, the root system of eggplant seedlings was used to identify SmTCP7a transcription factor binding sites during eggplant resistance to greening wilt. ChIP-seq analysis revealed insights into the transcriptional regulation mechanism and target genes. This study enhances our comprehension of the SmTCP7a regulatory mechanism in eggplant defense against Cyanobacteria.
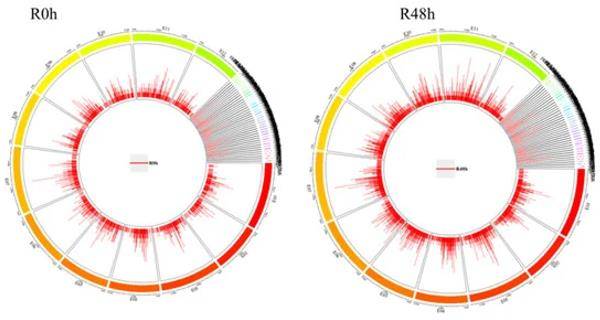 An overview of ChIP-Seq peak distribution in eggplant chromosomes for R0 h and R48 h. (Xiao et al., 2022)
An overview of ChIP-Seq peak distribution in eggplant chromosomes for R0 h and R48 h. (Xiao et al., 2022)
ChIP-seq has also been used in rice transcription factor research. The study reveals, for the first time, a novel mechanism by which the plant bZIP-type transcription factor APIP5, which has dual DNA- and RNA-binding activities, regulates cell death and defense responses in rice at the transcriptional and post-transcriptional levels by techniques such as ChIP-seq and ChIP-qPCR.
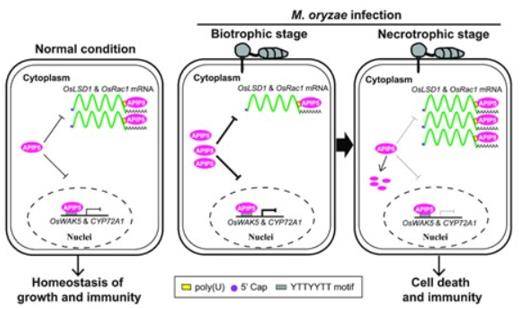 A working model illustrating the molecular mechanism of APIP5-mediated cell death and immunity to M. oryzae in rice. (Zhang et al., 2022)
A working model illustrating the molecular mechanism of APIP5-mediated cell death and immunity to M. oryzae in rice. (Zhang et al., 2022)
Conclusion
Transcription factor binding sites are the key to understanding gene regulation, and advancements in technology have paved the way for more accurate predictions and insights. The study of transcription factors and their binding sites not only sheds light on the intricate workings of gene expression but also has practical implications for fields like medicine and biotechnology. As we continue to explore the ever-expanding landscape of genomics, the role of transcription factors and their binding sites will undoubtedly remain at the forefront of biological research.
References:
- Xiao, Xi'ou, et al. "Genome-wide identification of binding sites for SmTCP7a transcription factors of eggplant during bacterial wilt resistance by ChIP-seq." International Journal of Molecular Sciences 23.12 (2022): 6844.
- Zhang, Fan, et al. "APIP5 functions as a transcription factor and an RNA-binding protein to modulate cell death and immunity in rice." Nucleic acids research 50.9 (2022): 5064-5079.


 Sample Submission Guidelines
Sample Submission Guidelines
