Genome-Wide Association Study: Introduction, Methods, and Application
Introduction to Genome-wide Association Studies
The National Institutes of Health defines a Genome-wide Association Study as "a study of common genetic variation across the entire human genome with the goal of identifying genetic associations with observable traits." Even though family linkage studies and studies with tens of thousands of gene-based SNPs assay genetic variation across the genome, the National Institutes of Health definition requires a decent density and selection of genetic markers to capture a large proportion of common variants in the study population, quantified in enough individuals to give sufficient power to detect variants of modest effect.
The focus of this discussion is on studies that attempt to assay at least 100 000 SNPs chosen to serve as proxies for the largest number of SNPs possible. A typical GWA study is divided into four sections: (1) a large number of people with the disease or trait of interest, as well as a suitable control group; (2) DNA isolation, genotyping, and data analysis to ensure high genotyping quality; (3) statistical tests for associations between SNPs that pass quality thresholds and disease/trait; and (4) replication of detected associations in a separate sample population or experimental examination of functional implications.
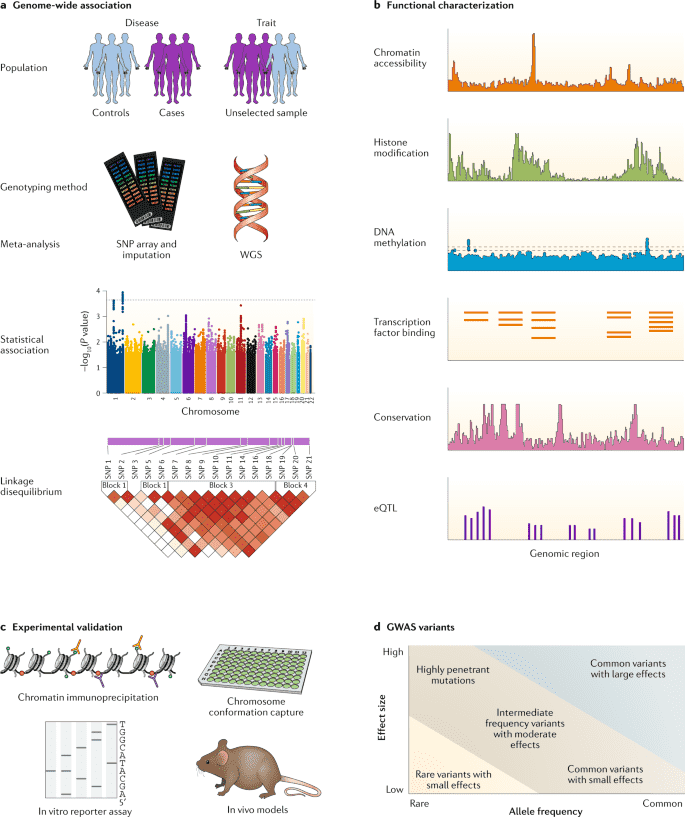
Figure 1. A quick view of Genome-Wide Association Study. (Tam, 2019)
Methods Used in genome-wide Association Studies
The case-control design, which compares two large groups of people, one healthy control group and one diseased case group, is the most common approach used in GWA studies. The majority of common known SNPs are genotyped in each group's members. The exact number of SNPs varies depending on genotyping technology, but it is usually one million or more. The allele frequency of each of these SNPs is then compared between the case and control groups to see if there is a significant difference. The odds ratio is the fundamental unit for reporting effect sizes in such situations. The odds ratio is the ratio of two odds, which in GWA studies are the odds of a case for people who have a specific allele and the odds of a case for people who do not have that same allele.
The assessment of quantitative phenotypic data, such as height or biomarker concentrations, or even gene expression, is a common alternative to case-control GWA studies. Alternative statistics for dominance and recessive penetrance patterns can also be used. Bioinformatics software such as SNPTEST and PLINK, which support many of these alternative statistics, are commonly used for calculations. The effect of individual SNPs is the focus of GWAS. Complex interactions between two or more SNPs, known as epistasis, may also play a role in complex diseases. Identifying statistically relevant interactions in GWAS data is computationally and statistically difficult due to the potentially infinite number of interactions.
Applications of Genome-wide Association Studies
GWA studies assay hundreds of thousands of single-nucleotide polymorphisms (SNPs) and link them to clinical conditions and measurable traits using high-throughput genotyping technologies. Since 2005, GWA studies have classified and replicated nearly 100 loci for as many as 40 common diseases and traits, many in genes not initially suspected of playing a role in the illness under study, and some in genomic areas containing no known genes. GWA studies are a significant step forward in the discovery of disease-causing genetic variants, but they also have significant drawbacks, such as the potential for false-positive and false-negative results, as well as biases related to study participant selection and genotyping errors. Although applications of GWA findings in prevention and treatment are currently being pursued, these studies primarily serve as a valuable discovery tool for analyzing genomic function and clarifying pathophysiologic mechanisms.
References:
- Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nature genetics. 2007 Oct;39(10).
- Zhang H, Wang Z, Wang S, Li H. Progress of genome wide association study in domestic animals. Journal of animal science and biotechnology. 2012 Dec;3(1).
- Pearson TA, Manolio TA. How to interpret a genome-wide association study. Jama. 2008 Mar 19;299(11).
- Tam V, Patel N, Turcotte M, et al. Benefits and limitations of genome-wide association studies. Nature Reviews Genetics. 2019 Aug;20(8).


 Sample Submission Guidelines
Sample Submission Guidelines
