Gene Expression Profiling Techniques: A Comprehensive Overview
In the realm of molecular biology, gaining insight into gene expression is of paramount significance for disentangling the elaborate processes that dictate cellular function. Techniques for profiling gene expression have been established as influential instruments that aid in decoding the multifaceted milieu of gene activity at the cellular level. In this scholarly communication, we endeavor to probe deep into the heart of gene expression profiling. Our focus will rest on shedding light on its foundational principles, methodologies, and utility in elucidating the complexities intertwined with cellular functionality and the onset of diseases.
What is Gene Expression?
Gene expression denotes the intricate progression wherein the genetic blueprint encapsulated within DNA undergoes conversion into functional gene products, namely proteins or RNA molecules. This meticulously orchestrated process entails a series of tightly regulated stages, comprising transcription, RNA processing, and translation. Within the dynamic milieu of cellular activity, a cell judiciously selects a subset of its genetic repertoire to express at any given moment, guided by internal cues and external stimuli. This discerning expression pattern ultimately dictates the cell's phenotype and function, underscoring the pivotal role of gene regulation in cellular physiology.
What is Gene Expression Profiling?
Gene expression profiling constitutes a methodological framework employed to concurrently assess the activity of myriad genes within a cellular or tissue specimen. Through quantification of gene expression levels, researchers can discern the molecular intricacies orchestrating diverse biological phenomena, encompassing cell differentiation, disease evolution, and drug reactivity. This analytical approach facilitates the identification of genes exhibiting differential expression patterns under distinct experimental settings, thereby furnishing a comprehensive portrayal of cellular dynamics.
Why Use Gene Expression Profiling?
Contemporary research fields encompassing developmental biology, oncology, pharmacology, and personalized medicine typically exploit the utilities offered by gene expression profiling. Such utilities offer an insight into gene regulation completely, potential disease biomarkers for both diagnosis and prognosis, and the discovery of therapeutic intervention points.
Crucially, by synergistically integrating other omics datasets - including but not limited to genomics and proteomics - with gene expression data, researchers have the capacity to achieve a holistic understanding of biological systems. This comprehensive overview facilitates the laying of foundational insights, helping to propel the boundaries of our current knowledge sphere.
Understanding Cellular Dynamics
Gene expression profiling empowers researchers to dissect the intricate dynamics governing cellular processes by scrutinizing alterations in gene expression patterns. For instance, delving into gene expression profiles throughout cellular differentiation unveils the molecular mechanisms propelling developmental pathways and fostering tissue specialization. This analytical approach affords profound insights into the orchestrated interplay of genetic elements orchestrating cellular fate determination, thereby enriching our understanding of fundamental biological phenomena.
Disease Diagnosis and Prognosis
In the realm of medical research, gene expression profiling harbors significant promise for the diagnosis and prognosis of diverse diseases, notably cancer. Through the scrutiny of gene expression patterns within diseased tissues, clinicians can discern molecular signatures linked to distinct disease subtypes, thereby facilitating precise diagnosis and informed therapeutic strategies. This analytical approach underscores the pivotal role of molecular characterization in tailoring personalized interventions, ultimately enhancing patient care and clinical outcomes.
Drug Discovery and Development
Molecular biology demonstrates the irreplaceable role gene expression profiling bears in advancing drug discovery and development. It is an impelling factor in scrutinizing therapeutic targets and investigating biomarkers with novel potential. Upon perturbing gene expression levels with drug treatments, the subsequent response underscores the efficacy of the treatment and reveals salient mechanisms of drug action. This innovative approach, consequently, furthers the identification and development of new drug candidates.
What Are Gene Expression Profiling Techniques
Given the substantial advances in molecular biology, gene expression profiling techniques have emerged as a fundamental component, offering researchers sophisticated instruments for deciphering the exquisite complexities of cellular operations. These methodologies are employed broadly across a wide array of research domains, spanning from developmental biology and cancer biology to pharmacology and personalized medicine.
Such techniques bestow upon researchers the capacity to illuminate gene regulatory networks, recognize biomarkers applicable to disease diagnosis and prognosis, and expose promising therapeutic avenues. Researchers are then enabled to integrate gene expression data with additional layers of omics information, such as genomic or proteomic data, to attain a holistic comprehension of biological systems. This integrated approach allows for a deeper understanding, promoting breakthroughs and aiding in the development of groundbreaking innovations in the life sciences.
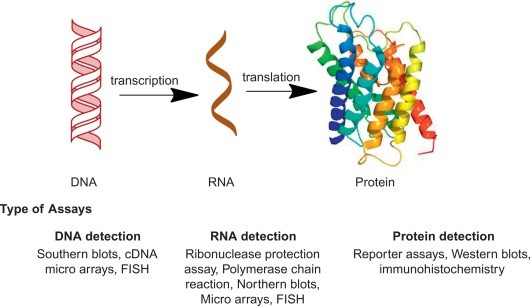 Central dogma of gene expression and list of biochemical assays used for their detection at transcription and translation level.
Central dogma of gene expression and list of biochemical assays used for their detection at transcription and translation level.
Here are some gene expression profiling techniques:
RNA Sequencing (RNA-Seq) Techniques for Gene Expression Profiling
RNA-Seq is undeniably a revolutionizing contribution to molecular biology, demystifying the profound labyrinth of gene expression profiling. This discourse explores an array of methodologies and applications underpinning RNA sequencing, underscoring its crucial role in unraveling the perplexing intricacies of the transcriptomic terrain and deepening our comprehension of the dynamism intrinsic to gene expression. Across a broad span of research disciplines, which notably include developmental biology, oncology, pharmacology, and personalized medicine, the application of these gene expression profiling methodologies has been influential. By facilitating the delineation of gene regulatory networks, the identification of disease diagnostic and prognostic biomarkers, and the discovery of promising therapeutic targets, RNA-Seq has been pivotal. Further, RNA-seq also demonstrates its powerful potential when coupled with other 'omics' data, such as genomics and proteomics, offering researchers an enriched, holistic insight into biological systems.
You may interested in
Unveiling Transcriptomic Complexity
Given the precision and breadth of insight provided by RNA sequencing methodologies, they have irrefutably revolutionized and enriched the field of gene expression analysis. Contrary to traditional microarray techniques, RNA-Seq exhibits heightened sensitivity and resolution, capable of detecting an assortment of RNA molecules. This includes mRNA transcripts, non-coding RNAs, as well as splice variants and fusion transcripts. The all-encompassing perspective offered by RNA-Seq elucidates the multifaceted intricacies underpinning gene expression regulation, thus enhancing our understanding of biological health and disease states.
Profiling Differential Gene Expression
A cornerstone application of RNA sequencing is the profiling of differential gene expression across diverse biological conditions. By quantifying mRNA abundance with high precision, RNA-Seq facilitates the identification of genes that are upregulated or downregulated in response to specific stimuli, treatments, or disease states. This information provides valuable insights into cellular pathways, regulatory networks, and pathophysiological mechanisms, paving the way for targeted therapeutic interventions and precision medicine approaches.
Deciphering Alternative Splicing Patterns
Alternate splicing serves as an essential mechanism for augmenting transcriptomic diversity and magnifying proteomic complexity. Through the application of RNA sequencing methodologies, detailed insights into alternate splicing behaviors become discernible, as they facilitate the characterization of exon utilization, splice site fluctuations, and transcript isoform prevalence. When coupled with exhaustive analysis, the dynamics of splicing implicated in RNA-Seq data reveal the subtle and intricate mechanisms involved in controlling gene expression at the post-transcriptional stage. Such insights afford a profound understanding of a variety of biological phenomena including but not limited to, the progression of developmental processes, the modulation of tissue-specific functionalities, and the intricate origins and perpetuation of disease pathogenesis.
Exploring Non-Coding RNA Landscapes
In the broader spectrum, beyond protein-coding genes, RNA sequencing opens a window to the exploration of the enigmatic world of non-coding RNAs, which encompasses microRNAs, long non-coding RNAs, and circular RNAs. These non-coding RNAs function as key orchestrators in the regulation of genes, the modulation of epigenetic dynamics, and contribute significantly to the understanding of disease pathophysiology. By charting the expression blueprints of these diverse non-coding RNA forms, RNA-Seq expounds their functional contributions, regulatory interplays, and potential diagnostic value. This greatly enhances our comprehension of RNA-mediated processes, both in health and disease phenotypes.
Leveraging Advanced Sequencing Technologies
The breakthrough in recent years regarding RNA sequencing technologies has extended the capabilities of RNA-Seq in gene expression profiling. Technological developments, such as single-cell RNA sequencing, long-read sequencing, and spatial transcriptomics, have ushered in a paradigm shift in transcriptomic analysis. This allows for the dissection of gene expression patterns with unparalleled resolution and granularity. These advanced technologies empower researchers to probe into cellular heterogeneity, developmental trajectories, and the progression of disease states, thus opening up unexplored fronts for potential discoveries and ground-breaking innovations within the sphere of genomics.
Microarray Analysis: Unraveling Gene Expression Patterns
Microarrays for gene expression serve as a high-throughput framework capable of quantifying the expression levels of thousands of genes in a concurrent manner. These microarrays comprise DNA probes that are immobilized onto a stationary substrate, thereby enabling the hybridization of RNA samples labelled with fluorescent tags. While these microarrays present cost-effective and scalable strategies in profiling gene expression, their inherent limitations include the inability to discern novel transcripts and a heightened vulnerability to ambient noise interference. Nevertheless, despite such limitations, microarray analysis continues to stand as a powerful methodology for large-scale studies on gene expression.
Quantitative PCR (qPCR): Precision in Gene Expression Quantification
On the other hand, the quantitative reverse transcription qRT-PCR persists as the exemplar technique for high specificity, sensitivity-based gene expression quantification. This method enables meticulous quantitation of mRNA abundances, proving immensely beneficial particularly for the verification of gene expression data gleaned from other profiling methods. Widely utilized in both clinical and research environments, qPCR - with its succinct and straightforward workflow - stands a pivotal role in molecular biology. Its ability to rapidly amplify and detect specific DNA sequences fosters applications spanning diagnostic, forensic and research pursuits.
Digital PCR: Precision in Gene Expression Quantification
More recently, digital PCR has emerged as a pioneering method for the precise and accurate quantification of gene expression. This innovative technique disperses a PCR reaction into thousands of independent reactions, with each one encapsulating an individual template molecule. The subsequent phase involves digitally enumerating the partitions that test positive or negative, thereby facilitating the accurate quantification of specific RNA transcript abundance. In comparison to traditional qPCR, digital PCR holds an edge in terms of augmented sensitivity, linearity, and repeatability, proving to be an indispensable tool for applications that call for precise quantification of gene expression levels.
Leveraging Single-Cell RNA Sequencing
The innovation of single-cell RNA sequencing (scRNA-Seq) has profoundly remodeled our comprehension of cellular diversity and transcriptional nuances at an individual cellular resolution. By creating transcriptomic portraits of distinct cells, scRNA-Seq pierces the veil of cellular heterogeneity, uncovering rare cellular populations and tracing developmental lineages within intricate tissue structures. This sophisticated methodology enables an unprecedented identification of specific cell types, lineage pathways, and regulatory mechanisms. Consequently, it has been instrumental in driving significant advances in fields such as developmental biology, immunology, and oncology.
Spatial Transcriptomics: Mapping Gene Expression in Tissues
Spatial transcriptomics is an emergent field that melds the potency of RNA sequencing with the precision of spatial resolution imaging methods. The resultant visualization allows researchers to extrapolate gene expression patterns within unprocessed tissue specimens. This pioneering methodology bestows researchers with the ability to spatially profile transcript distribution in tissue sections, offering a profound understanding of the spatial arrangement of gene expression within multifaceted biological constructs.
The potential applications of spatial transcriptomics are numerous, and broadly encompass the elucidation of tissue architecture, the probing of cellular interactions, and the identification of disease-associated spatial expression patterns. This technique is remarkable in that it integrates spatial information with transcriptional data, presenting a comprehensive perspective of dynamic gene expression set within the anatomical context of tissues. Therefore, it demarcates an exciting avenue for dissecting the complex orchestration of biological activities across multiple spatial scales.
NanoString Technology: High-Throughput Gene Expression Profiling
Implementing NanoString technology, researchers gain access to a high-capacity, multiplexed platform for accurate and sensitive quantification of gene expression levels. This innovative method employs molecular barcodes coupled with digital analytics, creating a precise measure of specific RNA transcript abundance in biological samples. The paramount benefit of NanoString assays lies in their capacity for parallel analysis of several hundred genes, despite having limited input materials. This characteristic renders it an ideal choice for examinations involving scant clinical samples or scarce cellular populations. Consequently, NanoString technology finds substantial utility in the exploration of biomarkers, molecular diagnostics, and the overarching field of translational research. This provides researchers critical insight into the mechanisms of diseases and the subsequent prospective therapeutic responses.
Ribosome Profiling: Decoding Translational Dynamics
Ribosome profiling, commonly referred to as Ribo-Seq, invites a fresh perspective on gene expression by capturing the 'footprint' of ribosomes across mRNA sequences. This high-resolution technique encompasses the comprehensive sequencing of mRNA fragments sheltered by ribosomes, thereby empowering researchers to decode the translational dynamics at play within the cell. Ribosome profiling paves the way for insights into aspects like translational efficacy, the degree of ribosome engagement, and the post-transcriptional modulation of gene expression. By enlisting this profiling method to chart actively translated mRNAs, researchers have the tools to demystify complex layers of genetic regulation and pinpoint genes under translational control in both health and disease states.
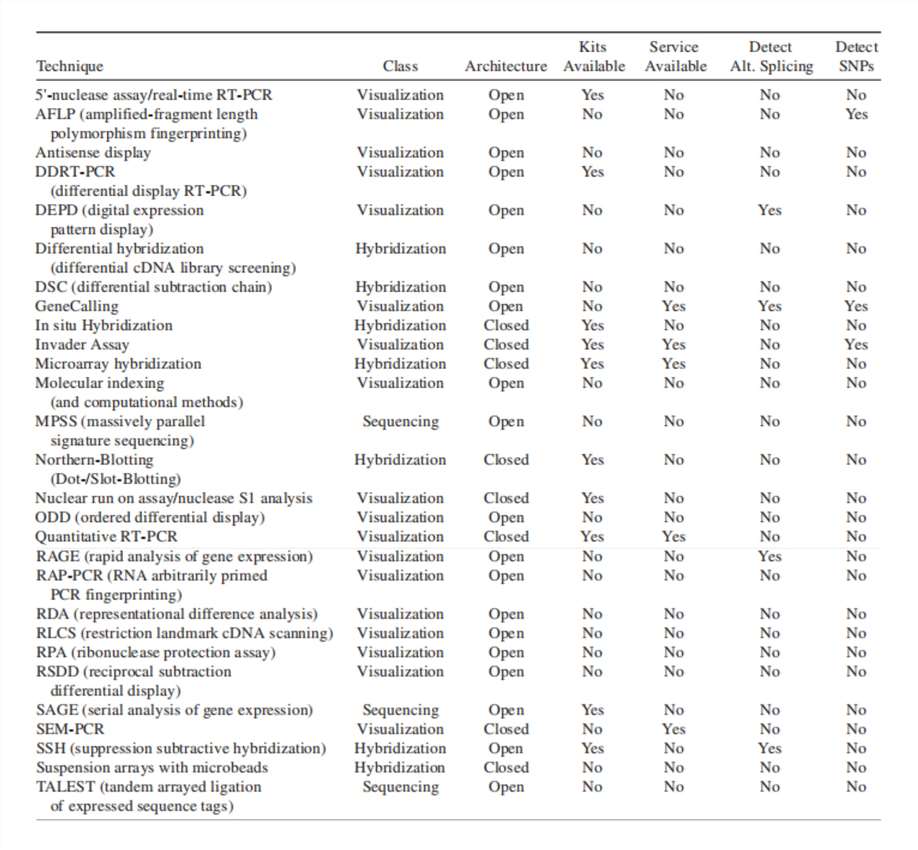 Other Gene Expression Profiling Methods (Richard A. Shimkets Methods in Molecular Biology, Vol. 258)
Other Gene Expression Profiling Methods (Richard A. Shimkets Methods in Molecular Biology, Vol. 258)
Conclusion: Empowering Precision in Gene Expression Profiling
Ultimately, RNA sequencing represents a pioneering advancement in gene expression profiling and provides exceptional insights into the intricate dynamics and regulations of cellular transcriptomes. Capitalizing on the versatility of RNA-Seq, scientists are now able to disentangle the intricacies of gene expression at an unprecedented resolution, sensitivity, and depth. CD Genomics continues its commitment towards the progression of RNA sequencing technologies and in doing so, provides innovative approaches for gene expression analysis—inescapable in powering scientific discovery and precision medicine across a diverse spectrum of research disciplines.
References:
- Liang M, Cowley AW, Greene AS. High throughput gene expression profiling: a molecular approach to integrative physiology. J Physiol. 2004
- Ahmed, R. Narain, in Polymers and Nanomaterials for Gene Therapy, 2016


 Sample Submission Guidelines
Sample Submission Guidelines
