Complex Telomere-Related Variants in Cancer Genomes through Genome Sequencing
Telomeres, specialized genome structures comprising short, poly-repeated non-transcribed sequences (TTAGGG) and associated binding proteins, undergo pivotal changes in structure and location during the evolution of cancer genomes. Historically, the limitations of short-read Next-Generation Sequencing (NGS) technologies hindered the characterization of repetitive arrays within long telomeres, impeding our understanding of critical alterations in cancer. The advent of long-read sequencing technologies, particularly the highly precise PacBio HiFi sequencing, has revolutionized our ability to accurately assess telomere repeat-containing structures in cancer and cancer cell lines.
A recent publication has emerged, presenting an in-depth analysis of telomere repeat-containing structures in cancer and cancer cell lines. This analysis utilizes a combination of long-read and short-read sequencing technologies, including PacBio HiFi and Nanopore, to provide a comprehensive and precise evaluation.
Telomere Dynamics in Cancer Unraveled by Long-Read Sequencing
The in-depth analysis through long-read sequencing has unearthed novel insights into telomeric repeat sequences, particularly those occurring ectopically in the standard orientation. These sequences appear to be extensive and limitless, indicative of neotelomeric additions. For instance, examination of sequencing data identified a potential ectopic telomeric repeat sequence site neighboring the chrX:103,320,553 sequence in the U2-OS osteosarcoma cell line. This site exhibited a minimum of seven tandem (TTAGGG) repeats, accompanied by a discernible reduction in sequencing coverage at the corresponding chromosomal location—a hallmark of telomeric repeat presence.
In stark contrast, the PacBio HiFi and Oxford Nanopore long-read sequencing datasets provided a more comprehensive view, revealing long telomeric repeats spanning approximately 3-10kb in the canonical direction within this region. The observed variation in telomere length across read lengths suggests a potential association with the loss of active telomeric sequences post-DNA replication or ongoing telomere maintenance processes.
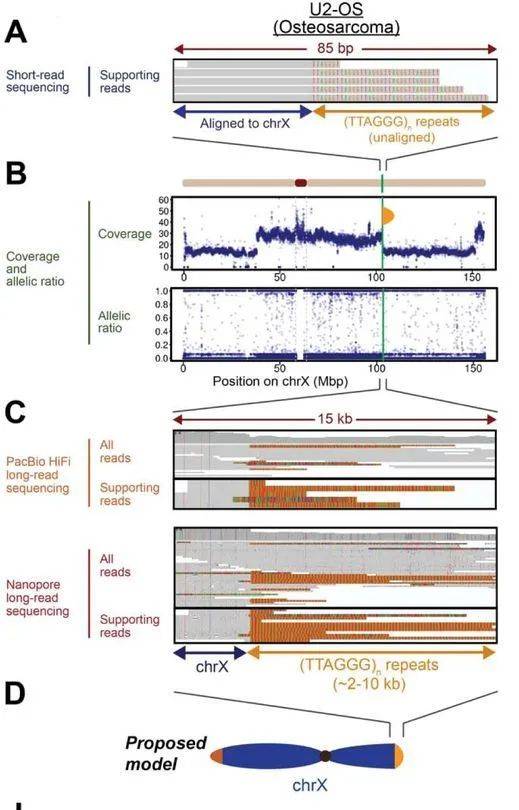 Neotelomeres in cancer genomes revealed by long-read genome sequencing. (Tan et al., 2023)
Neotelomeres in cancer genomes revealed by long-read genome sequencing. (Tan et al., 2023)
Long-Read Long Sequencing Unearths Chromosome Arm Fusion with Telomeric Repeat Sequences
In the pursuit of understanding sites hosting ectopic telomeric repeat sequences, the researchers made a striking discovery—these repeats were not only present but also exhibited a reverse orientation relative to the breakpoints. The application of long-read sequencing revealed that these loci predominantly signify chromosome arm fusion events.
A detailed examination of the candidate locus region, employing PacBio HiFi and Nanopore long-read sequencing, highlighted the presence of approximately 650 base pairs of reverse (CCCTAA) repeat sequences following the breakpoints. Additionally, 5-8 kilobases of sequences on the chr22q subtelomeres were observed. The coherent narrative emerges from a single long read encapsulating the entire event, suggesting that the reverse (CCCTAA)n repeat sequence originated from the fusion of the chr22q arm with its short telomere to an intrachromosomal site.
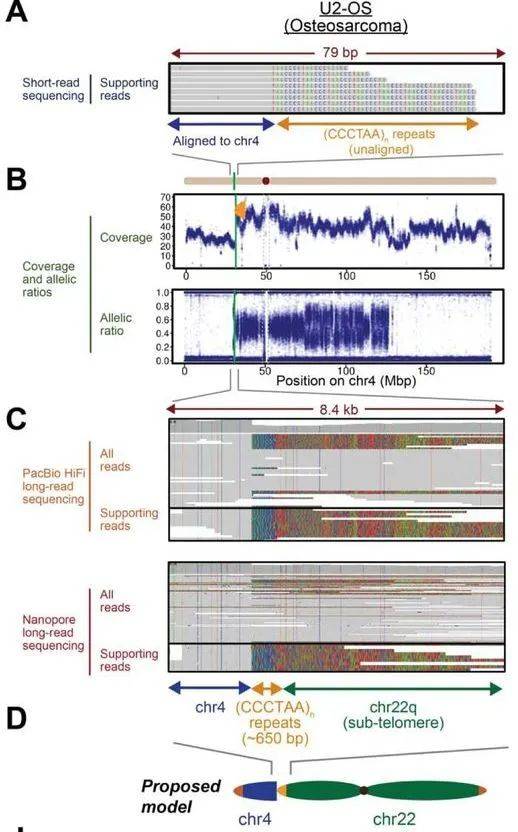 Chromosomal arm fusions in cancer genomes revealed by long-read genome sequencing. (Tan et al., 2023)
Chromosomal arm fusions in cancer genomes revealed by long-read genome sequencing. (Tan et al., 2023)
Disruption of Protein-Coding Genes by New Telomeres and Chromosome Arm Fusion Events
The introduction of telomeric insertions carries the inherent risk of disrupting crucial genes, including those governing tumor suppression, thereby inducing consequential functional ramifications. A notable illustration is the identification of a neotelomere (16 kb) within the first intron of the PTPN2 gene through long-read sequencing. This insertion led to abnormalities in the first exon and promoter region of the gene associated with immunotherapy response.
Moreover, the study brought to light chromosomal fusion events that instigated disruptions in genes, exemplified by occurrences that resulted in the loss of over half of the 5' region of the KLF15 and FOXN3 genes. These findings underscore the potential far-reaching impact of new telomeres and chromosome arm fusion events on the integrity and functionality of protein-coding genes, including those pivotal for regulating critical cellular processes.
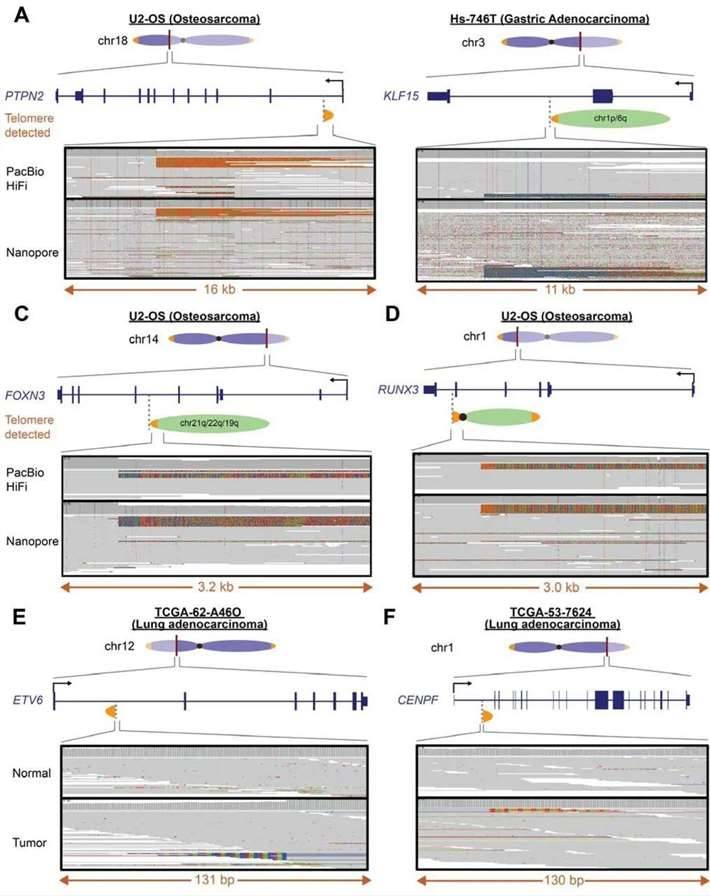 Neotelomeres and chromosomal arm fusion events disrupt protein coding genes in cancer cell lines and patient samples. (Tan et al., 2023)
Neotelomeres and chromosomal arm fusion events disrupt protein coding genes in cancer cell lines and patient samples. (Tan et al., 2023)
Reference:
- Tan, Kar-Tong, et al. "Neotelomeres and Telomere-Spanning Chromosomal Arm Fusions in Cancer Genomes Revealed by Long-Read Sequencing." bioRxiv (2023): 2023-11.


 Sample Submission Guidelines
Sample Submission Guidelines
