Fostering Tumor Heterogeneity: The Impact of Cyclic Extrachromosomal DNA (ecDNA)
What Is Cyclic Extrachromosomal DNA (ecDNA)?
Since the 1960s, the presence of cyclic extrachromosomal DNA (ecDNA) has been documented in isolated tumors and tumor-derived cells. Characterized as a circular, unfilamented chromatin body with variable lengths spanning from tens of kilobases to tens of megabase pairs (Mbp), cyclic ecDNA emerges as a pivotal factor contributing to intra-tumor heterogeneity. Its involvement in tumorigenesis, tumor evolution, and the development of drug resistance underscores its significance.
Cyclic ecDNA & Cancer
Cyclic ecDNA plays a prominent role as a prevalent form of high-copy oncogene amplification, serving as a prognostic biomarker across various tumor types. Notably, medulloblastoma featured prominently in the initial case reports detailing ecDNA. Unfortunately, the current therapeutic landscape for medulloblastoma lacks highly effective targeted molecular therapies. The existing standard of care carries substantial risks, including cognitive impairment, neurological damage, and the potential for secondary malignancies.
Medulloblastoma is stratified into four major molecular subgroups: WNT, SHH, type 3, and type 4. Prognostically, tumors with MYC activation in type 3 and those with TP53 mutations in SHH subtypes exhibit particularly dismal outcomes. This underscores the urgent need for innovative therapeutic approaches in managing medulloblastoma, given the challenges associated with current treatment modalities and the severity of prognosis within specific molecular subgroups.
ecDNA Exacerbates Oncogenic Impact in Medulloblastoma
In the investigation, 102 instances of extrachromosomal DNA (ecDNA) sequences were identified in tumor samples obtained from 82 individuals, constituting 18% of the 468 patients studied. Tumors with ecDNA positivity (ecDNA+) were distributed among patients as follows: WNT, 0 out of 22 total; SHH, 30 out of 112 (27%); type 3, 19 out of 107 (18%); and type 4, 26 out of 181 (14%). Remarkably, SHH subgroup tumors exhibited a higher likelihood of containing ecDNA compared to other medulloblastoma subgroups.
Within this cohort, ecDNA-amplified genes observed in two or more samples encompassed well-established or suspected medulloblastoma oncogenes, including MYC, MYCN, MYCL, TERT, GLI2, CCND2, PPM1D (WIP1), and ACVR2B. Additionally, genes associated with DNA repair mechanisms (RAD51AP1 and RAD21) and those encoding inhibitors of the TP53 pathway (PPM1D28 and CDK6) were among the identified ecDNA-amplified candidates. These findings shed light on the complex genetic landscape of medulloblastoma, emphasizing the potential therapeutic implications associated with ecDNA-amplified oncogenes in this malignancy.
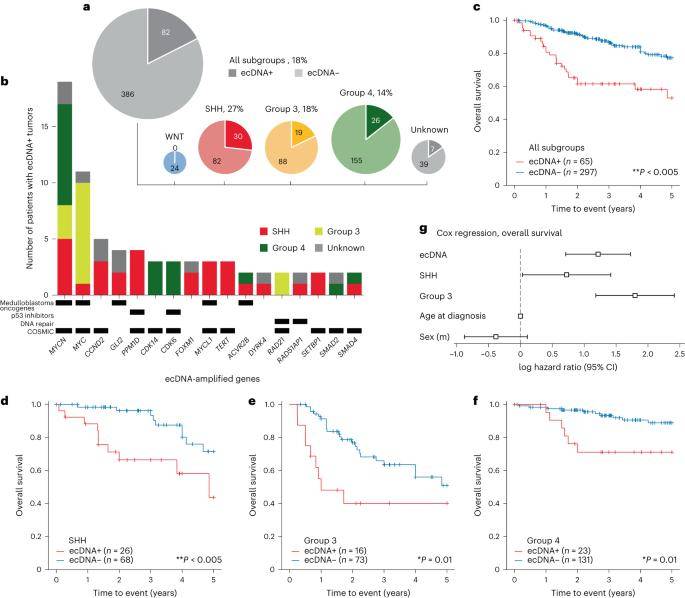 The landscape of ecDNA in medulloblastoma patient tumors. (Chapman et al., 2023)
The landscape of ecDNA in medulloblastoma patient tumors. (Chapman et al., 2023)
ecDNA as an Indicator of Adverse Prognosis in Medulloblastoma
Patients harboring tumors with extrachromosomal DNA (ecDNA) exhibited significantly diminished five-year overall survival and progression-free survival in comparison to those with ecDNA-negative (ecDNA-) tumors. Notably, overall survival was notably lower among patients with ecDNA+ tumors within the SHH, type 3, and type 4 subgroups (P < 0.05). To further assess the prognostic significance of ecDNA, Cox proportional hazards regression analysis was conducted, revealing that individuals with ecDNA+ tumors faced a heightened prognostic risk and increased mortality compared to their counterparts with ecDNA- tumors (P < 0.005). These findings underscore the potential of ecDNA as a robust prognostic marker in medulloblastoma, highlighting its association with poorer clinical outcomes.
Association of TP53 Mutations with ecDNA in SHH Medulloblastoma
The tumor suppressor protein p53, encoded by TP53, plays a pivotal role in regulating DNA damage perception, cell cycle arrest, and apoptosis, frequently encountering somatic mutations and pathogenic germline variants in SHH medulloblastoma. To explore the potential link between TP53 mutations and the presence of extrachromosomal DNA (ecDNA), researchers scrutinized the somatic and germline TP53 mutation status in 92 cases of SHH medulloblastoma.
Notably, TP53 mutations were more prevalent in ecDNA+ SHH tumors compared to their ecDNA- SHH counterparts. However, this correlation was not evident in other medulloblastoma subtypes or the entire cohort, suggesting a subtype-specific functional relationship between TP53 mutations and ecDNA, particularly within the SHH subtype. Cox regression analysis revealed that the impact of ecDNA on survival couldn't be solely attributed to TP53 mutations. While causation cannot be definitively inferred from these data alone, these analyses pinpoint TP53 mutations and ecDNA as clinically relevant biomarkers for highly aggressive SHH medulloblastoma tumors.
Coexistence of Diverse ecDNA Profiles in Select Medulloblastomas
Analysis of 16 medulloblastoma cases revealed the presence of multiple distinct extrachromosomal DNA (ecDNA) sequences. Notably, a primary SHH medulloblastoma tumor exhibiting a heterozygous TP53 mutation (RCMB56-ht) was selected for the establishment of a patient-derived orthotopic xenograft mouse model (RCMB56-pdx). In-depth whole-genome sequencing (WGS) and optical genome mapping (OGM) were employed for genome assembly, confirming the existence of a circular amplicon (amp1) comprising three DNA fragments on chromosome 1. Additionally, a continuous chromatin amplicon (amp2) consisting of 21 fragments on chromosomes 7 and 17 was validated.
To ascertain the co-occurrence of these distinct ecDNA profiles within the same cells, multichannel fluorescence in situ hybridization (FISH) imaging of the same markers in interphase cells was conducted. The presence of different fluorescent spots for each gene within the nucleus of the same cell indicated that each copy of the amplified gene resided on a separate chromatin body. These findings underscore the intricate nature of ecDNA diversity within medulloblastomas, providing valuable insights into the complex genomic landscape of these tumors.
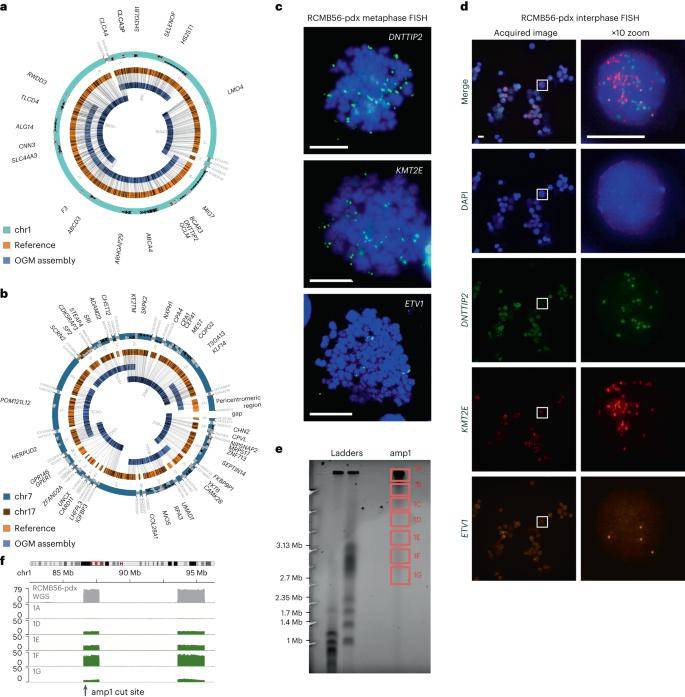 Distinct high-copy extrachromosomal amplifications coexist in a SHH medulloblastoma tumor. (Chapman et al., 2023)
Distinct high-copy extrachromosomal amplifications coexist in a SHH medulloblastoma tumor. (Chapman et al., 2023)
Heterogeneity in ecDNA Copy Numbers Across Medulloblastoma
In this investigation, researchers established an automated image analysis pipeline to assess the copy number distribution per cell in interphase fluorescence in situ hybridization (FISH) microscopy, aiming to quantify the heterogeneity of extrachromosomal DNA (ecDNA) in medulloblastoma. The mean and variance of the estimated copy numbers per cell for all ecDNA amplification marker genes were markedly higher compared to the ecDNA cell line COLO320-HSR, which encompasses the chromosomally amplified MYC locus. This aligns with the observed high copy number heterogeneity in human cancer cell lines featuring ecDNA.
Notably, in each analyzed primary tumor, only 22%-41% of cells exhibited an ecDNA-amplified copy number greater than 5. Insights gleaned from quantitative FISH microscopy and multichromosomal single-cell sequencing indicate that a subset of cells within ecDNA+ medulloblastoma tumors harbors high-copy ecDNAs. Furthermore, these cells display highly variable single or multiple extrachromosomal amplified copy numbers. These findings underscore the intricate and dynamic nature of ecDNA copy number variations within the cellular landscape of medulloblastomas.
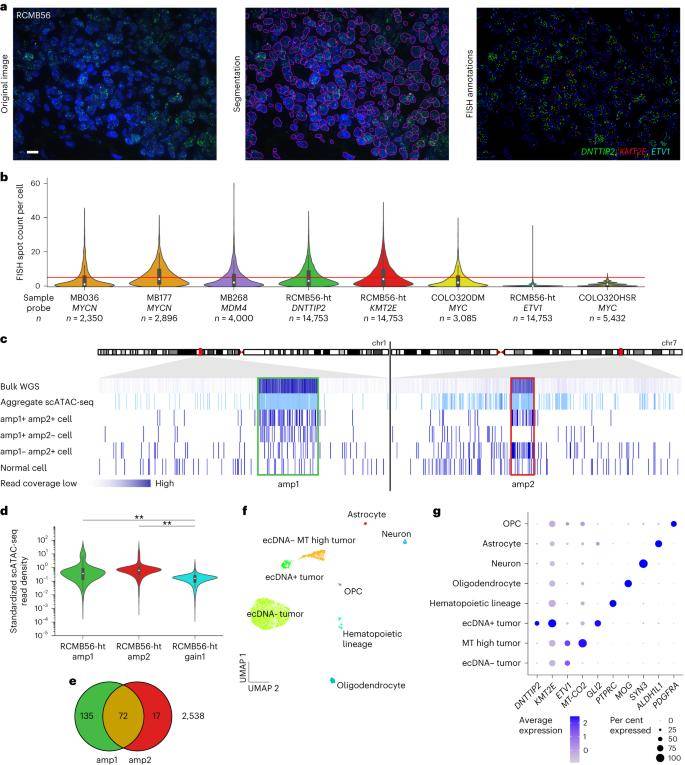 Single-cell analysis reveals a distinct tumor cell population with high-copy ecDNA amplification. (Chapman et al., 2023)
Single-cell analysis reveals a distinct tumor cell population with high-copy ecDNA amplification. (Chapman et al., 2023)
Unique Transcriptomic Profiles in ecDNA+ Cells
Utilizing the weighted nearest neighbor algorithm for clustering, a majority of ecDNA+ cells were segregated into distinct clusters characterized by unique transcriptomic and epigenetic profiles. Notably, cells within the ecDNA+ cluster exhibited significant overexpression of the amp1 and amp2 marker genes DNT IP 2 (Wilcoxon rank-sum test, q < 0.001) and KMT2E (q < 0.001). Furthermore, GLI2 (q < 0.001) was markedly upregulated in the ecDNA+ cell cluster compared to other tumor and normal cells. The transcriptional activity of ecDNA-amplified genes demonstrated a positive correlation with increasing ecDNA copy numbers.
To contextualize these findings, normal cell types were identified through cluster-specific expression of known marker genes. Single-nucleus RNA sequencing (snRNA-seq) genomic copy number estimation corroborated that normal cells maintained a stable genome, contrasting with tumor cell clusters exhibiting a diverse array of copy number alterations. These results highlight the distinct molecular landscape of ecDNA+ cells, emphasizing their unique transcriptional signatures and underlying genomic alterations compared to both tumor and normal cells.
ecDNA Orchestrates Oncogene Placement within an Aberrant Gene Regulatory Landscape
Given the profound genomic rearrangements linked to medulloblastoma ecDNA, the study delved into whether abnormal DNA interactions between co-amplified oncogenes and enhancers occur within the context of circular ecDNA. To explore this, accessible chromatin from 25 medulloblastoma tumors was subjected to analysis through ATAC-seq, while chromatin interactions from 17 tumors were investigated using chromatin conformation capture (Hi-C).
In half of the scrutinized ecDNA+ tumors (D458, MB106, MB268, and RCMB56), compelling evidence of aberrant chromatin interactions on ecDNA was identified, spanning structurally variable breakpoints. These interactions facilitated the juxtaposition of accessible motifs and co-amplified genes originating from distal genomic regions. Hi-C data further revealed that the MYC promoter exhibited interactions with chromosomes 8 and co-amplified regulatory elements on chromosome 14.
These findings underscore the prevalence of abnormal enhancer-promoter interactions arising from structural rearrangements of ecDNA in medulloblastoma tumors. The study sheds light on the intricate regulatory dynamics orchestrated by ecDNA, providing valuable insights into the molecular underpinnings of medulloblastoma pathogenesis.
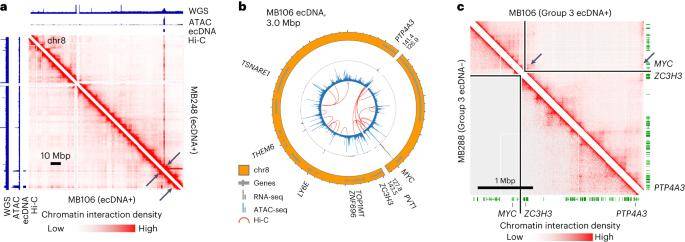 Chromatin interactions with MYC are rewired in a Group 3 medulloblastoma. (Chapman et al., 2023)
Chromatin interactions with MYC are rewired in a Group 3 medulloblastoma. (Chapman et al., 2023)
Enhancer-Mediated Regulation of Oncogene Transcription through ecDNA Amplification
A comprehensive CRISPRi amplification screen was conducted in the type 3 medulloblastoma cell line D458, employing 32,530 sgRNA sequences targeting 645 accessible sites on extrachromosomal DNA (ecDNA). The same screening approach was applied to the type 3 cell line D283, characterized by MYC amplification within a 55 Mbp uniformly stained region of chromosome 8q. While the MYC promoter proved essential in both cell lines, the screen pinpointed six functional elements that specifically impeded D458 proliferation following 21 days of CRISPRi inhibition.
Additional findings from the inhibition screen underscored that, although MYC amplification propels proliferation across all type 3 medulloblastoma cell lines, the relative significance of co-amplified genes and cis-regulatory elements varied based on the genomic structure of the amplicon. This highlights the nuanced interplay between amplified oncogenes and associated regulatory elements, shedding light on the complex landscape of oncogene transcriptional control orchestrated by ecDNA amplification in medulloblastoma.
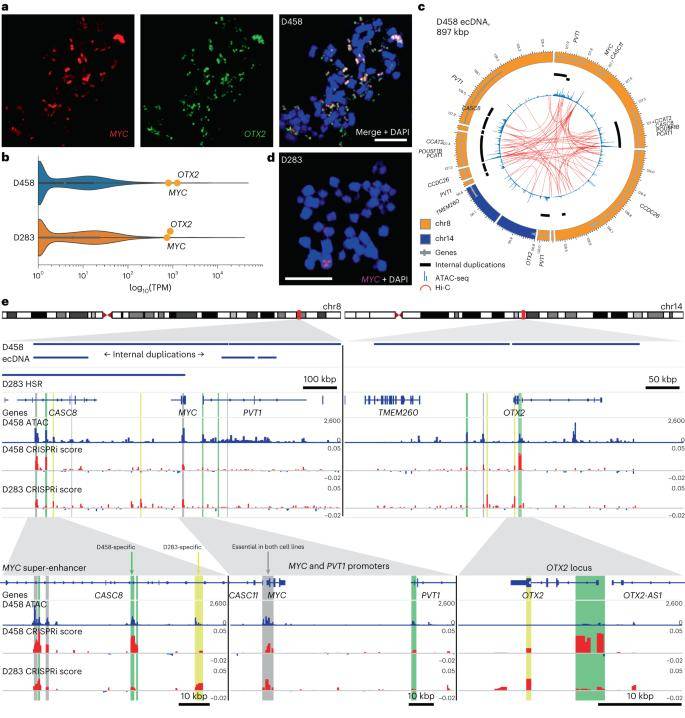 Enhancer rewiring in medulloblastoma ecDNA affects cell proliferation. (Chapman et al., 2023)
Enhancer rewiring in medulloblastoma ecDNA affects cell proliferation. (Chapman et al., 2023)
Reference:
- Chapman, O.S., Luebeck, J., Sridhar, S. et al. Circular extrachromosomal DNA promotes tumor heterogeneity in high-risk medulloblastoma. Nat Genet 55, 2189–2199 (2023).


 Sample Submission Guidelines
Sample Submission Guidelines
