What is B-Cell Receptor Repertoire Sequencing?
What is BCR-seq?
B cell receptor sequencing (BCR-seq) represents an advanced and rapid method for the exhaustive detection of target-amplified B-cell receptors (BCRs) through the integration of high-throughput sequencing technology and bioinformatics. By scrutinizing the abundance of each sequence, BCR-seq unveils the intricacies of B-cell-mediated humoral immune responses and the dynamic diversity of immunoglobulin alterations. This cutting-edge technology enables a thorough exploration of the underlying mechanisms governing the immune system's ability to respond and adapt, opening avenues for a deeper understanding of immune system dynamics.
CD Genomics also provides TCR Sequencing service based on next-generation sequencing and long-read sequencing.
BCR Structure
The B-cell receptor (BCR) serves as a membrane-bound immunoglobulin (Smlg), playing a pivotal role in antigen recognition by B cells. Structurally, the BCR is a tetramer, comprising two heavy chains (H) and two light chains (L), where the light chain can be either κ or λ. The H chain is intricately composed of four gene fragments: 65-100 types of variable regions (VH), two types of diversity regions (DH), six types of joining regions (JH), and constant regions (CH).
On the other hand, the L chain encompasses three distinct gene fragments, including variable regions (VH), joining regions (JH), and constant regions (CH). Specifically, the L chain consists of variable regions (VH), joining regions (JH), and constant regions (CH), with each region contributing to the overall functionality of the BCR. This meticulous arrangement ensures the BCR's ability to recognize a diverse array of antigens, forming a critical component in the intricate machinery of the immune response.
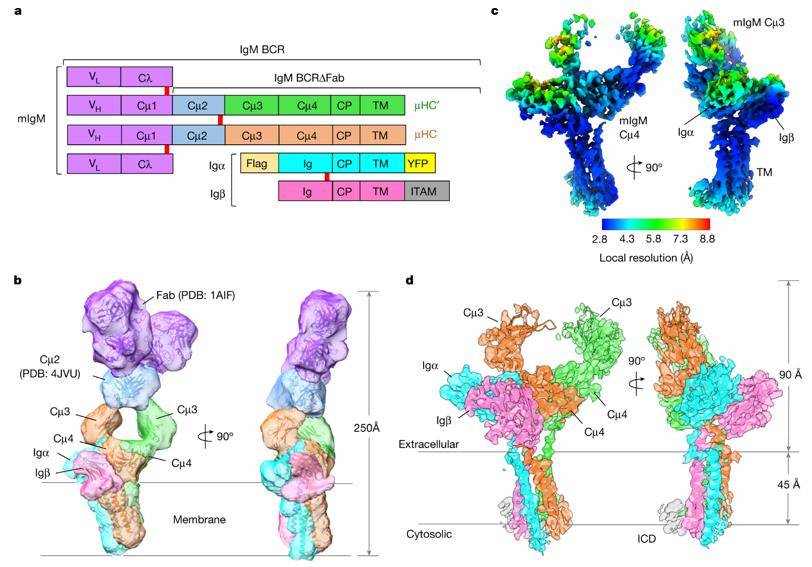 Cryo-EM maps of the IgM BCR. (Dong et al., 2022)
Cryo-EM maps of the IgM BCR. (Dong et al., 2022)
BCR Diversity
- Diversity Arising from Gene Fragment Rearrangement (V, D, and J)
The variable (V) regions of both heavy (H) and light (L) chains play a crucial role in BCR diversity. Comprising three amino acids each, these regions exhibit high variability, forming complementarity-determining regions (CDRs) — CDR1, CDR2, and CDR3. All three CDRs actively participate in antigen recognition, collectively determining the BCR's antigenic specificity. Notably, the gene encoding CDR3, positioned at the junction of light chain V and J or heavy chain V, D, and J fragments, enhances BCR diversity by incorporating nucleotide insertions or splicing during V(D)J rearrangements and V-D and D-J junctions.
- Diversity Stemming from Nucleotide Sequence Combinations
BCR diversity is further enriched through the combination of numerous V-region gene fragments, both in light and heavy chains. Additionally, the diverse combinations of V, D, and J genes contribute significantly. During rearrangement, each gene can only be taken in one fragment, accentuating the overall diversity of BCRs.
- Diversity Induced by High-Frequency Somatic Cell Mutations
Mature B cells within peripheral lymphoid organ germinal centers undergo increased mutation frequency in the rearranged V region gene (CDR3) upon stimulation by antibodies. Referred to as high-frequency mutations in somatic cells, these mutations, predominantly point mutations, occur non-randomly. This process contributes to the dynamic adaptability of BCRs, enhancing their ability to respond to a wide array of antigens.
The Workflow of B Cell Receptor (BCR) Sequencing
- Isolation and B Cell Capture
B cells are meticulously isolated from blood or tissue samples, and their RNA is extracted.
- Reverse Transcription to cDNA
The extracted RNA undergoes reverse transcription, transforming it into complementary DNA (cDNA).
- Receptor Library Preparation
Using the cDNA as a template, a receptor library is meticulously crafted. Specific primers, strategically designed with markers upstream and downstream based on gene characteristics, are employed. Following PCR amplification, a comprehensive receptor library is established. All resulting products, marked for identification, are poised for subsequent reaction sequencing.
- High-Throughput Sequencing
Employing state-of-the-art high-throughput sequencing technologies, the obtained cDNA sequences are meticulously recorded.
- Bioinformatics Data Processing
The acquired sequencing data undergoes a sophisticated bioinformatics analysis. This encompasses meticulous filtering of sequencing data, sequence splicing, VDJ sequence comparison, and ultimately, the determination and translation of sequence structures.
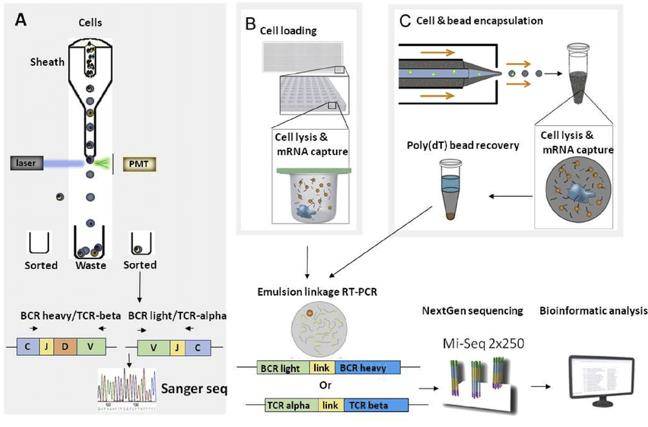 Approaches for BCR and TCR repertoire sequencing. (Seah et al., 2018)
Approaches for BCR and TCR repertoire sequencing. (Seah et al., 2018)
Applications of BCR Sequencing
- Unveiling Pathogenesis and Early Disease Monitoring
BCR sequencing serves as a powerful tool for delving into the intricacies of disease pathogenesis and enables early monitoring and diagnosis.
- Dynamic Monitoring of BCR CDR3 Receptor Library for Drug Efficacy
The dynamic monitoring of the BCR CDR3 receptor library provides a valuable avenue for observing the therapeutic effectiveness of drugs.
- Correlation of B-Cell Receptor Gene Loci with Disease Occurrence
Investigating the correlation between B-cell receptor gene loci and the onset and progression of specific diseases offers vital insights for early monitoring and understanding disease pathogenesis.
- Tumor Homology Analysis
BCR sequencing plays a pivotal role in analyzing tumor homology within an organism, contributing to our understanding of cancer biology and progression.
- Screening Somatic Cell Mutations in Primary Immunodeficiencies
It facilitates the screening of somatic cell mutations, particularly in patients with primary immunodeficiencies such as isolated IgA deficiency, aiding in personalized medical approaches.
- Tolerance Assessment in Transplant Rejection
BCR sequencing contributes to the assessment of tolerance in transplant rejection scenarios. It aids in predicting rejection occurrences and guides timely and effective intervention strategies.
References:
- Dong, Ying, et al. "Structural principles of B cell antigen receptor assembly." Nature 612.7938 (2022): 156-161.
- Seah, Yu Fen Samantha, Hongxing Hu, and Christoph A. Merten. "Microfluidic single-cell technology in immunology and antibody screening." Molecular aspects of medicine 59 (2018): 47-61.


 Sample Submission Guidelines
Sample Submission Guidelines
