What Are Antibiotic Resistance Genes (ARGs)?
A resistance gene denotes a nucleotide sequence (DNA fragment) responsible for encoding a trait that confers drug resistance. Similar to other genetic materials, these genes undergo replication during bacterial division and proliferation. These antibiotic-resistant genes may reside on the bacterial chromosome or on plasmids external to the chromosome. Notably, resistance genes on plasmids can be transmitted among strains of the same bacterial species or even different species through mechanisms such as splicing, transformation, and transduction.
In 2017, the World Health Organization (WHO) identified 12 particularly menacing "superbugs," underscoring the global emergence of bacterial antibiotic resistance as a critical public health concern.
Advancements in sequencing technologies and the evolution of bioinformatics have led to the widespread adoption of high-throughput sequencing techniques in the realm of bacterial antibiotic resistance detection. Noteworthy methods include whole genome sequencing (WGS), metagenomic sequencing (mNGS), and microbial single-cell transcriptome sequencing, also known as microbial single-cell RNA sequencing.
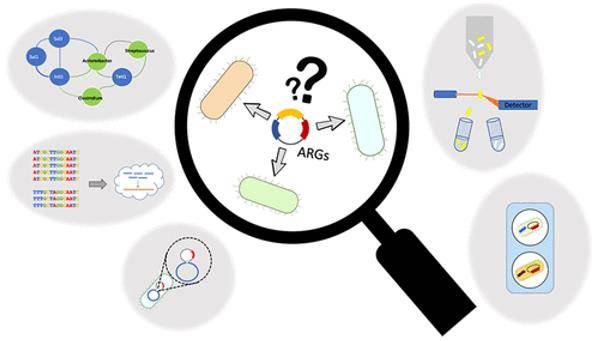 Antibiotic resistance genes (ARGs) detection. (Rice et al., 2020)
Antibiotic resistance genes (ARGs) detection. (Rice et al., 2020)
The Different Types of Resistance Genes
Resistance genes, based on the antibiotics that induce resistance, can be categorized into several types. These include tetracycline resistance genes (tet), sulfonamide resistance genes (sul), β-lactamase resistance genes (bla), macrolide resistance genes (erm), aminoglycoside resistance genes (aac), fluoroquinolone resistance genes (fca), colistin resistance genes (mcr), vancomycin resistance genes (van), and multidrug resistance genes (mdr).
The global dissemination of drug-resistant genes in the environment has raised significant concerns. Primary reservoirs for drug-resistant strains and genes are found in the intestinal tracts and feces of livestock and poultry, facilitating the widespread transmission of these genes. Mobile genetic elements (MGEs) play a pivotal role in this dissemination, encompassing crucial entities like insertion sequences, transposons, integrons, plasmids, and integrative splicing elements.
Horizontal gene transfer serves as a key mechanism for the transmission of drug-resistant genes, occurring through three common genetic processes:
- Splicing (Plasmid-Mediated): Involving the transfer of genetic material through plasmids.
- Natural Transformation (Bacterial Uptake of DNA from the Environment): Where bacteria take up DNA directly from their surroundings.
- Transduction (Mediated by Phage): Facilitated by bacteriophages, which act as carriers of genetic material between bacteria.
Understanding these mechanisms is essential for addressing the challenge of antibiotic-resistant gene proliferation and devising effective strategies to mitigate their impact on public health and the environment.
Mechanisms of Antibiotic Resistance
Antibiotic Resistance Genes (ARGs) encoding antibiotic resistance is an intrinsic phenomenon predating human influence. Bacteria exhibit resistance through diverse mechanisms, encompassing antibiotic efflux, impermeability of the cell membrane, modification of antibiotic targets, and the synthesis of antibiotic hydrolases. Notably, aminoglycosides, characterized by numerous macromolecules featuring exposed hydroxyl and amide groups, are particularly susceptible to modification, resulting in the manifestation of high levels of resistance.
Recent advancements have unveiled novel resistance mechanisms, including the ABC-F protein's role in antibiotic competition, mutations that incapacitate essential genes, and the fostering of resistance in challenging environmental conditions. These insights deepen our understanding of the dynamic and evolving landscape of antibiotic resistance, emphasizing the intricate interplay between bacteria and their response to selective pressures.
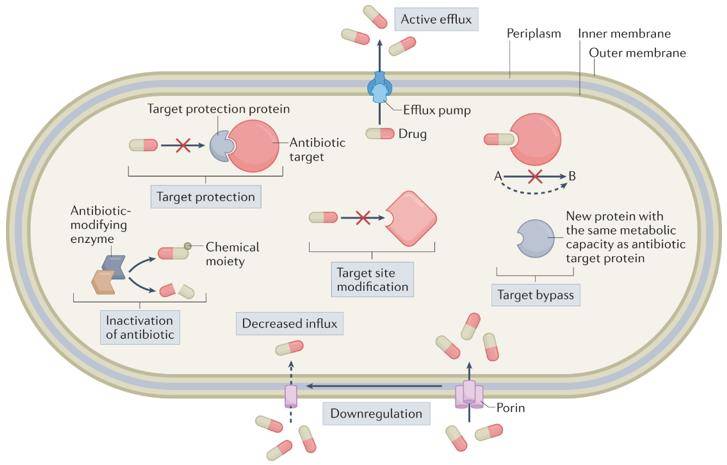 Overview of the molecular mechanisms of antibiotic resistance. (Darby et al., 2023)
Overview of the molecular mechanisms of antibiotic resistance. (Darby et al., 2023)
Bacterial Resistance Gene Detection Based on High-Throughput Sequencing
WGS is the comprehensive process of deciphering the entire genetic sequence of a given species. In the realm of bacterial drug resistance detection, WGS emerges as a powerful tool, offering a thorough profile of drug resistance genes within bacteria. It goes beyond mere identification, enabling the prediction of drug resistance phenotypes by analyzing the intricacies of drug resistance genes and unraveling the associated resistance mechanisms. Numerous studies have undertaken comparisons between WGS and phenotypic AST for bacterial drug resistance detection, consistently demonstrating the precision of WGS in predicting bacterial drug resistance.
Moreover, WGS plays a crucial role in elucidating antibiotic resistance genes present in both chromosomes and plasmids within the genomes of antibiotic-resistant bacteria. This knowledge contributes significantly to understanding the dynamics behind the movement and dissemination of resistance genes. Additionally, WGS serves as a valuable tool for monitoring outbreaks and epidemics of antibiotic-resistant strains. Despite these advantages, it's important to note that WGS has certain drawbacks, including a lengthier testing period, higher costs, and the requirement for pure bacterial cultures. These factors contribute to the heightened demands associated with the identification and isolation of clinical bacteria using this technique.
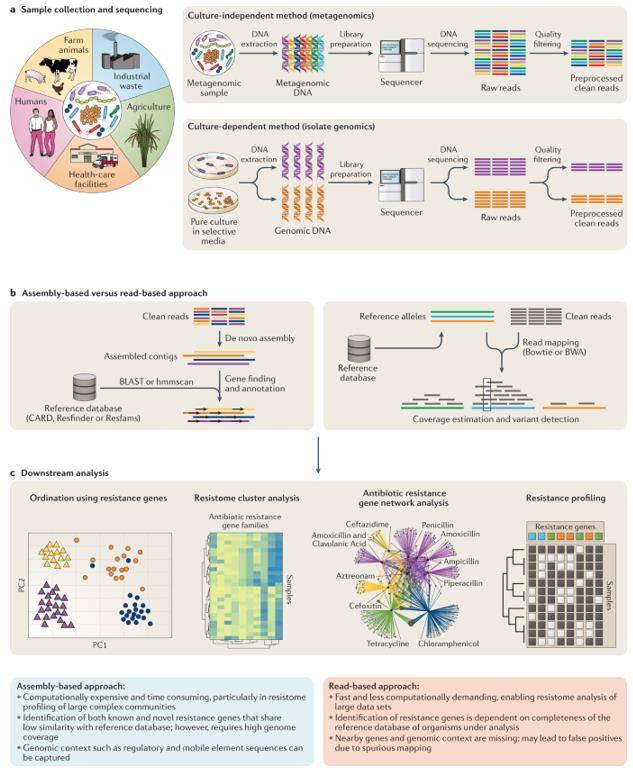 Assembly versus read mapping. (Boolchandani et al., 2019)
Assembly versus read mapping. (Boolchandani et al., 2019)
mNGS is a cutting-edge approach designed to unveil the genomic information of microbial populations within specific environmental samples through high-throughput sequencing technology. Leveraging bioinformatics analysis methods, mNGS facilitates pathogen identification, microbiome analysis, host interaction studies, and prediction of bacterial drug resistance. In the realm of clinical bacterial drug resistance gene detection, mNGS emerges as a powerful tool capable of diagnosing infections caused by unknown pathogens and uncovering associated drug resistance genes. This information serves as a valuable reference for guiding the judicious use of antibiotics in clinical settings.
In a notable study, Wang et al. utilized mNGS technology to identify Klebsiella pneumoniae in culture-negative lung tissue samples from an immunocompromised patient with severe pneumonia. The analysis further revealed resistance genes such as blaSHV-12, blaKPC-2, blaTEM-1, and blaCTX-M-65. Additionally, mNGS proves instrumental in the detection of novel resistance genes, who identified 11 previously unknown antibiotic resistance genes from soil samples. These included novel ampicillin resistance genes, gentamicin resistance genes, chloramphenicol resistance genes, and meclobutanil resistance genes.
One key advantage of mNGS lies in its unbiased detection of antibiotic resistance genes directly from original samples, enabling the search for novel antibiotic resistance genes alongside the screening of existing ones. However, it is essential to acknowledge that this technology comes with inherent challenges, including high costs, complexity, and the absence of a standardized and automated analysis process. These factors present hurdles for the large-scale screening of bacterial drug resistance genes.
- Single-cell RNA Sequencing (scRNA-seq)
Single-cell RNA sequencing (scRNA-seq) revolutionizes our understanding of cellular populations by decoding the transcriptome of individual cells, unraveling population variances, and exposing evolutionary relationships. In recent years, microbial single-cell RNA sequencing has emerged as an advanced technology, employing scRNA-seq principles specifically on microorganisms, such as bacterial cells.
The development of MscRNA-seq faces challenges due to the low mRNA content of microorganisms, the distinctive cell wall and membrane features of certain bacteria, and the absence of mRNA polyadenylation. These technical barriers have historically impeded the progress of microbial scRNA-seq. However, Georg Seelig's research team at the University of Washington pioneered a breakthrough by employing microSPLiT sequencing of microbial single-cell transcriptomes using split-pool ligation-based transcriptome sequencing (SPLiT-seq) technology on bacteria.
Microbial SPLiT-seq represents a significant advancement, being the first cost-effective, high-throughput method for single-cell transcriptome sequencing of bacteria. MicroSPLiT, utilizing barcode-labeled RNA sources from bacterial cells, can analyze tens of thousands of bacterial cells in a single experiment.
As microbial scRNA-seq technology evolves, it successfully addresses challenges such as low mRNA content, cellular diversity, and complex cell wall structures. Its application extends to mining the transcriptome of drug-resistant genes in clinical superbugs. By comprehensively understanding the expression and transfer of drug-resistant genes, microbial scRNA-seq holds crucial significance for advancing the detection and screening of clinical multi-drug resistance.
Summary
Traditional methods of bacterial resistance detection offer valuable phenotypic insights into bacterial resistance but are hampered by prolonged culture and characterization times, and they fall short in identifying resistance genes. Emerging technologies rooted in physical, chemical, and molecular biology methods present advantages such as a shorter detection cycle and heightened sensitivity in bacterial drug resistance detection. However, these methods face limitations in detecting unknown resistance genes and mutated genes.
The advent of high-throughput sequencing-based detection technologies serves as a complementary approach, bridging the gap between classical phenotypic detection and molecular methods. This advanced technology excels in accurately identifying bacterial resistance genes, uncovering new resistance genes, and pinpointing mutation sites. Looking ahead, the integration of phenotypic and genotypic detection in bacterial drug resistance will be pivotal for controlling drug-resistant bacterial infections and managing epidemics. This holistic approach provides innovative technical support for the prevention and monitoring of related infectious diseases.
References:
- Rice, Eric W., et al. "Determining hosts of antibiotic resistance genes: a review of methodological advances." Environmental Science & Technology Letters 7.5 (2020): 282-291.
- Boolchandani, Manish, Alaric W. D'Souza, and Gautam Dantas. "Sequencing-based methods and resources to study antimicrobial resistance." Nature Reviews Genetics 20.6 (2019): 356-370.
- Darby, Elizabeth M., et al. "Molecular mechanisms of antibiotic resistance revisited." Nature Reviews Microbiology 21.5 (2023): 280-295.


 Sample Submission Guidelines
Sample Submission Guidelines
