Next Generation Sequencing for COVID-19
What is COVID-19?
COVID-19, formally designated as Coronavirus Disease 2019, is an exceptionally infectious respiratory disease incited by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This virus is a member of the Coronaviridae family, a group that incorporates numerous strains with the capacity to infect both humans and animals. The initial eruption of COVID-19 was pinpointed in December 2019, and it rapidly proliferated globally, culminating in an expansive pandemic. The disease principally disseminates via respiratory droplets produced when an infected individual coughs, sneezes, or speaks, thus rendering human-to-human transmission the primary channel for the virus's propagation.
COVID-19 manifests an extensive assortment of symptoms ranging from mild to severe. The most frequently encountered early indicators encompass fever, parched cough, and fatigue. It can, however, provoke more profound complications such as acute respiratory distress syndrome (ARDS), pneumonia, organ failure, and potentially lead to fatal outcomes—predominantly in older adults and individuals with pre-existing health conditions. Emerging research also posits that COVID-19's impact extends beyond the respiratory tract to encompass a wider range of organ systems, including cardiovascular, gastrointestinal, renal, and neurological.
Various stratagems have been employed in the bid to counteract the catastrophic reach of the COVID-19 pandemic, including extensive testing, tracking contacts of confirmed cases, implementation of quarantine methods, advocating social distancing, as well as the formulation of vaccines and therapeutics. A comprehensive understanding of the genetic composition and evolutionary trajectory of SARS-CoV-2 is imperative in crafting efficacious control procedures and confining the virus' spread. Next-generation sequencing (NGS) has ascended to prominence as an instrumental technique in unravelling SARS-CoV-2's genetic blueprint; this thereby enables researchers to monitor its transmission, scrutinize emergent mutations, and formulate context-specific interventions.
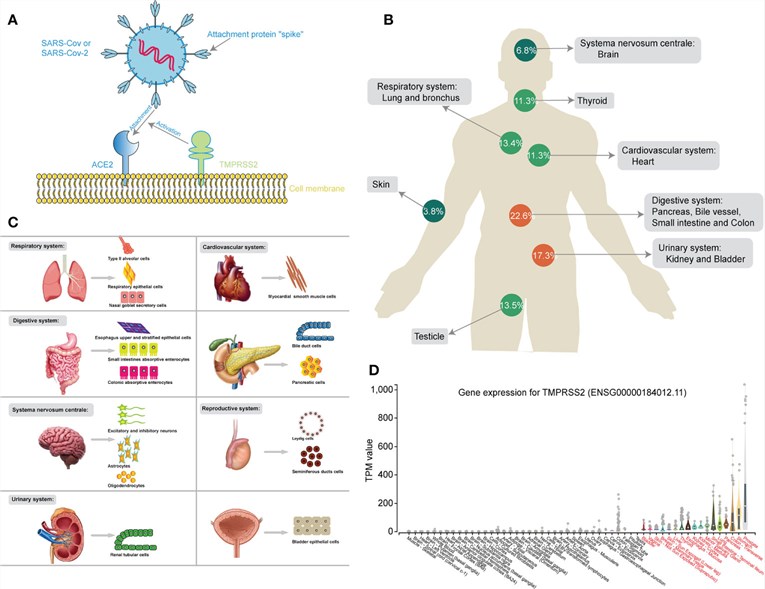 Mechanism of SARS-CoV-2
Mechanism of SARS-CoV-2
What is COVID-19 Next Generation Sequencing?
COVID-19 NGS entails the utilization of cutting-edge genomic sequencing methodologies for the scrutiny of the genetic composition of the severe acute respiratory SARS-CoV-2, the causative agent of COVID-19. NGS methodologies afford expedited and high-capacity sequencing of viral genomes, furnishing invaluable insights into the genetic landscape, evolutionary trajectories, and transmission dynamics of SARS-CoV-2. By comprehensively sequencing the entirety of the viral genome, investigators can discern mutational signatures, delineate viral lineages, and probe the genesis of emergent variants.
The advent of NGS technology has engendered a paradigm shift in our ability to investigate infectious maladies, COVID-19 included, by furnishing a panoramic vista of the viral genomic architecture with unparalleled celerity and precision. Diverging from conventional sequencing modalities, which entail protracted timelines and laborious workflows, NGS empowers researchers to concurrently sequence myriad DNA fragments, thereby markedly truncating the temporal and fiscal outlays entailed in genomic data acquisition. This scalability and efficacy render NGS an indispensable asset for scrutinizing viral epidemics and crafting precision-targeted countermeasures.
You may interested in
What are Sequencing Methods for COVID-19
Sequencing methodologies for COVID-19 encompass a spectrum of techniques facilitating the interrogation of the viral genome, thereby elucidating its genetic architecture, mutational landscape, and transmission kinetics. These methodologies are pivotal in deciphering the virus's pathophysiological underpinnings, informing public health interventions, and refining diagnostic and therapeutic modalities.
Amplicon Sequencing
Within the realm of COVID-19 NGS, targeted amplicon sequencing emerges as a cornerstone technique, characterized by the selective amplification and sequencing of designated segments of the viral genome. This method entails the design of primers targeting conserved regions within the SARS-CoV-2 genome, facilitating the amplification and subsequent sequencing of pivotal segments, including but not limited to the spike protein gene or the RNA-dependent RNA polymerase (RdRp) gene. Leveraging targeted amplicon sequencing affords rapid and cost-efficient identification of viral variants and mutations, thereby furnishing critical insights into the evolutionary dynamics and epidemiological nuances of the virus.
Hybrid Capture Sequencing
Hybrid capture sequencing encompasses the capture and subsequent sequencing of the complete viral genome through the utilization of complementary probes meticulously tailored to bind to defined genomic segments. This methodology ensures exhaustive coverage of the viral genome, facilitating the discernment of novel variants and mutations. Of particular significance for surveillance and epidemiological inquiries, hybrid capture sequencing furnishes intricate insights into the genetic heterogeneity of the virus and its transmission kinetics.
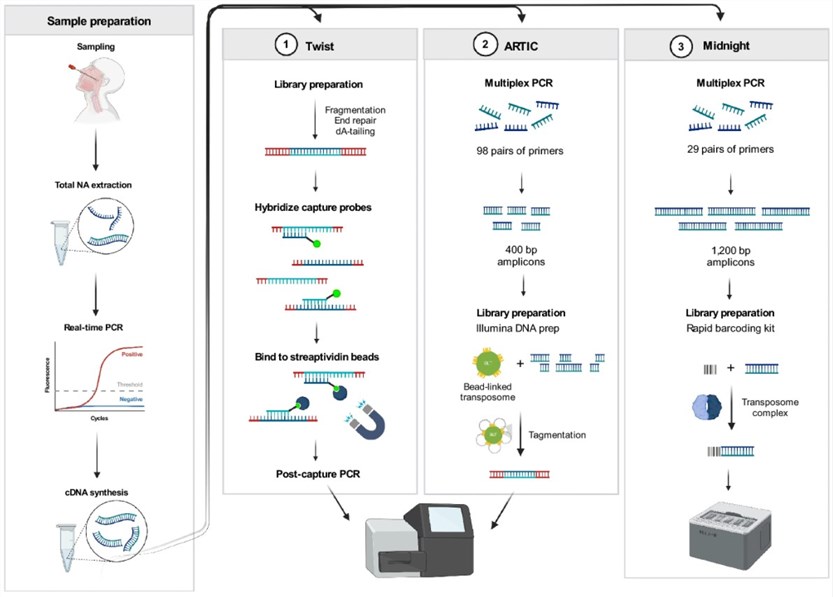 SARS-CoV-2 whole genome sequencing using tiled amplicon enrichment and bait hybridization
SARS-CoV-2 whole genome sequencing using tiled amplicon enrichment and bait hybridization
Shotgun Metagenomics
Shotgun metagenomic sequencing has emerged as a pivotal tool for unbiased detection and comprehensive characterization of viral pathogens within intricate samples, such as clinical specimens or environmental matrices. Within the context of COVID-19, metagenomic sequencing facilitates the discernment of SARS-CoV-2 within patient samples, concurrently unveiling the presence of other respiratory pathogens. By virtue of sequencing all nucleic acids inherent to a given sample, metagenomic methodologies afford the capability to uncover co-infections, delineate microbial interplay, and identify potential confounding variables influencing disease progression. Notably, metagenomic sequencing has proven instrumental in scrutinizing the intricate dynamics of SARS-CoV-2 transmission and scrutinizing outbreaks across diverse epidemiological landscapes.
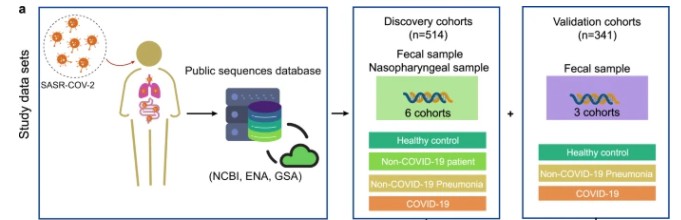 shotgun metagenomic sequencing of COVID-19
shotgun metagenomic sequencing of COVID-19
Whole Genome Sequencing
Viral whole Genome Sequencing, a branch of NGS, has proven to be a vital element in comprehending COVID-19's global distribution and genetic diversity. Uninhibited by preferential gene targets, WGS undertakes the task of sequencing the full viral genome, encompassing both coding and non-coding regions. This comprehensive analysis avails the detection of unprecedented mutations, facilitates tracing the proliferation of specific viral lineages, and offers insights into any genomic characteristics associated with either virulence or transmissibility. Acknowledging its significant contributions, it becomes evident that WGS has been instrumental in unearthing the emergence and global propagation of new SARS-CoV-2 variants including Alpha, Beta, Gamma, and Delta.
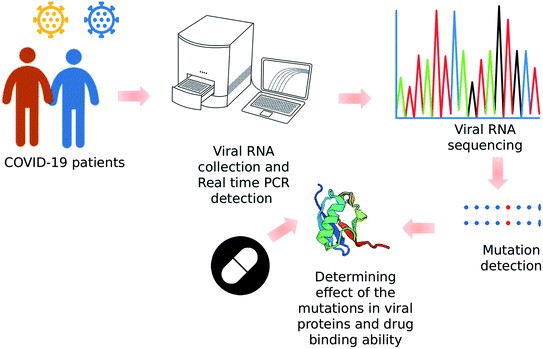 Whole genome sequencing for revealing the point mutations of SARS-CoV-2 genome
Whole genome sequencing for revealing the point mutations of SARS-CoV-2 genome
Long-read Sequencing Platforms
Long-read sequencing platforms, typified by Oxford Nanopore Technologies, have induced radical transformations in the approach used for SARS-CoV-2 genomic analysis. These innovative platforms outperform traditional short-read sequencing systems, generating protracted DNA reads which streamline the construction of complete viral genomes in a singular, continuous string of data. The exhaustive genomic data, procured via long-read sequencing, permits the execution of multiple applications pivotal for decoding the intricacies of the SARS-CoV-2 virus.
In evaluating SARS-CoV-2, long-read sequencing exhibits multiple superiorities. One significant gain is the generation of extensive reads, capacitating researchers to traverse multifaceted genomic terrains, inclusive of repetitive sequences and structural variants. This functionality emerges as especially pertinent in delineating the genomic framework of SARS-CoV-2, which manifests structural intricacies exemplified by RNA secondary structures and sequence disparities. More so, long-read sequencing platforms simplify the process of isolating viral subtypes and mutants, equipping us with profound insights into the routes of viral evolution and transmission modalities.
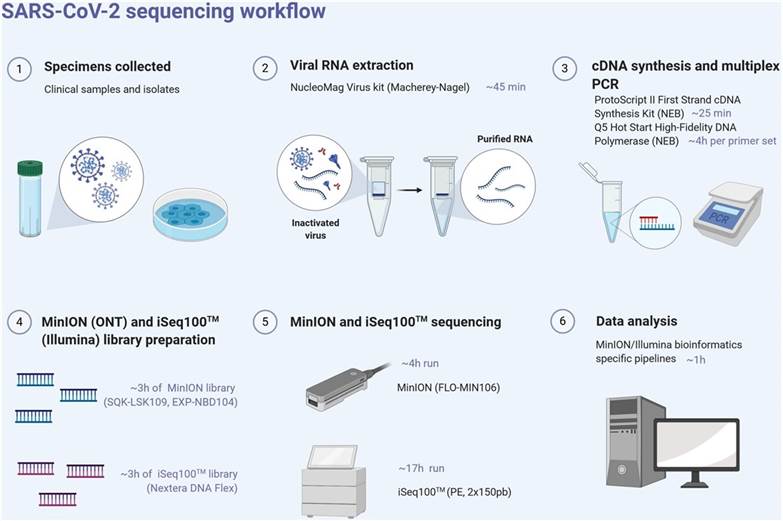 SARS-CoV-2 next-generation sequencing workflow (MinION), from the sample to sequence analysis.
SARS-CoV-2 next-generation sequencing workflow (MinION), from the sample to sequence analysis.
Comparative Analysis of Coronavirus NGS Methods
| Aspect | Targeted Amplicon Sequencing | Hybrid Capture | Shotgun Metagenomics | Ultra-High Throughput Sequencing | Whole Genome Sequencing |
| Testing Needs | Requires specific primers targeting known regions of the viral genome | Requires probes targeting specific viral sequences | Unbiased detection of all genetic material in a sample | Requires Oxford Nanopore platforms | Requires high-throughput sequencing platforms |
| Speed & Turnaround Time | Rapid turnaround time; Typically within hours to days | Moderate turnaround time; Typically days to weeks | Longer turnaround time; Typically days to weeks | Rapid turnaround time; Typically within hours to days | Rapid turnaround time; Typically within hours to days |
| Scalable & Cost-Effective | Scalable and cost-effective for targeted regions | Scalable but may be costly due to probe design and synthesis | Scalable but may require significant computational resources | Highly scalable but may be costly due to equipment and reagents | Highly scalable but may be costly due to equipment and reagents |
| Identify Novel Pathogens | Limited ability to identify novel pathogens outside of targeted regions | Limited ability to identify novel pathogens beyond captured sequences | Ability to identify novel pathogens based on sequence data | Ability to identify novel pathogens based on sequence data | Ability to identify novel pathogens based on sequence data |
| Track Transmission | Limited ability to track transmission | Limited ability to track transmission | Ability to track transmission based on genomic similarities | Ability to track transmission based on genomic similarities | Ability to track transmission based on genomic similarities |
| Detect Mutations | High sensitivity for detecting known mutations | High sensitivity for detecting known mutations | Ability to detect novel mutations throughout the viral genome | Ability to detect novel mutations throughout the viral genome | Ability to detect novel mutations throughout the viral genome |
| Identify Co-Infections & Complex Disease | Limited ability to identify co-infections or complex disease | Limited ability to identify co-infections or complex disease | Ability to identify co-infections or complex disease | Ability to identify co-infections or complex disease | Ability to identify co-infections or complex disease |
| Detect Antimicrobial Resistance | Not applicable for detecting antimicrobial resistance | Not applicable for detecting antimicrobial resistance | Limited ability to detect antimicrobial resistance without specific analysis | Limited ability to detect antimicrobial resistance without specific analysis | Limited ability to detect antimicrobial resistance without specific analysis |
COVID-19 NGS Application
Insights into Genomic Surveillance and Epidemiology
NGS technology functions as an invaluable resource in terms of tracing the evolutionary trajectory and global dispersion of SARS-CoV-2. Employing virus genomes harvested from patient samples, scientists have the capability to spot the advent of new variants whilst identifying genetic mutations tethered to alterations in virulence, transmissibility, and immune evasion. The key contributions of CD Genomics in the large-scale genomic surveillance projects have significantly enriched our comprehension of COVID-19 epidemiology and modality of transmission.
Enabling Precision in Diagnostic Testing
The employment of NGS-centric methodologies powerfully enables the diagnostic precision for COVID-19, as well as facilitating the detection of genetic variants that may influence viral infectivity and drug responsiveness. Sequencing viral RNA derivatives from patient extracts allows NGS to pinpoint SARS-CoV-2 with high precision and to distinguish it from other respiratory pathogens. Moreover, NGS can detect viral mutations allied with drug resistance, generating critical information to orientate future treatment strategies and direct antiviral drug discovery.
Steering Vaccine Development
The swift evolution of efficacious vaccines against COVID-19 is heavily rooted in a comprehensive understanding of the viral genome and its antigenic characteristics. NGS delivers an enhanced capacity to typify SARS-CoV-2 antigens and distinguish immunogenic epitopes that kindle protective immunological responses. By sequencing the viral isolates harvested from disparate geographic regions and patient cohorts, investigators can devise vaccine designs that target conserved sectors of the viral genome, thereby diminishing the likelihood of vaccine-escape mutations.
Inspiring Public Health Interventions
NGS-driven genomic surveillance serves as a pivotal linchpin in informing strategic public health interventions aimed at curtailing the dissemination of COVID-19 and mitigating its repercussions among vulnerable populations. By delineating transmission clusters and tracing the origins of outbreaks, NGS empowers the implementation of targeted quarantine measures, contact tracing endeavors, and isolation protocols. Moreover, the wealth of genomic data furnished by NGS facilitates the formulation of evidence-based guidelines concerning vaccine allocation, social distancing protocols, and travel restrictions, thereby augmenting the effectiveness of pandemic management strategies.
FAQ
A: How are viruses sequenced?
Q: NGS platforms facilitate the deconstruction of viral genomes from biological specimens, such as blood or respiratory secretions, into discrete, manageable units for analysis. The process commences with the extraction of the viral genetic material from the sample. Subsequent fragmentation of the nucleotides ensues, yielding smaller constituent sequences. These fragments are subsequently fed into high-throughput sequencing platforms, concurrently generating an array of miniature DNA sequences numbering in the millions. Utilising an array of computational biology tools, these fragmented sequences are reassembled into extensive contiguous sequences, colloquially termed as "reads". These reads collectively epitomise the virus's complete genetic blueprint. It is this sophisticated approach that permits the rapid and precise elucidation of the viral genome, a critical factor in the study of viral evolution, the dynamics of transmission and for generating potent diagnostic protocols and efficacious vaccines.
A: How did genome sequencing play an important role during the COVID-19 pandemic?
Q: The inauguration of genomic sequencing was paramount in managing several aspects of the COVID-19 pandemic. Most significantly, it fostered the swift discernment and elucidation of the novel coronavirus SARS-CoV-2, the pathogen associated with the COVID-19 disease. Through the genomic sequencing of viral samples accrued from patients, the scientific community was able to reveal the genetic composition of the virus encompassing its origination, variations, and evolutionary association with other coronaviruses. The gleaned information was vital for interpreting the transference dynamics of the pathogen, surveilling its worldwide propagation, and promoting the formulation of effective diagnostic assays and vaccines.
Moreover, genomic sequencing substantiated the identification of emerging SARS-CoV-2 variants as the course of the pandemic unfolded. By performing genetic sequencing on viral specimens from diverse temporal and geographical points, researchers succeeded in distinguishing mutations embedded within the viral genome which yielded certain advantages such as enhanced transmissibility or resistance to antibodies. The timely monitoring of these variants equipped public health establishments with the data required for executing specific interventions to modulate their distribution and to adjust vaccination strategies accordingly.
Additionally, the employment of genomic sequencing supplied researchers with the means to scrutinize the host's molecular-level response to SARS-CoV-2 invasion. Through the detailed analysis of the genetic blueprints of those infected, scientists acquired an understanding of the mechanisms driving disease severity, immune responses, and potential therapeutic targets. The insights derived from these studies profoundly contributed to the genesis of treatment protocols and personalized therapeutic strategies in managing COVID-19 patients.
References:
- Goswami, C., Sheldon, M., Bixby, C. et al. Identification of SARS-CoV-2 variants using viral sequencing for the Centers for Disease Control and Prevention genomic surveillance program. BMC Infect Dis 22, 404 (2022).
- Chen X, Kang Y, Luo J, Pang K, Xu X, Wu J, Li X, Jin S. Next-Generation Sequencing Reveals the Progression of COVID-19. Front Cell Infect Microbiol. 2021
- John G, Sahajpal NS, Mondal AK, Ananth S, Williams C, Chaubey A, Rojiani AM, Kolhe R. Next-Generation Sequencing (NGS) in COVID-19: A Tool for SARS-CoV-2 Diagnosis, Monitoring New Strains and Phylodynamic Modeling in Molecular Epidemiology. Curr Issues Mol Biol. 2021
- Hossain MU, Ahammad I, Bhattacharjee A, Chowdhury ZM, Hossain Emon MT, Chandra Das K, Keya CA, Salimullah M. Whole genome sequencing for revealing the point mutations of SARS-CoV-2 genome in Bangladeshi isolates and their structural effects on viral proteins. RSC Adv. 2021
- Koskela von Sydow, A., Lindqvist, C.M., Asghar, N. et al. Comparison of SARS-CoV-2 whole genome sequencing using tiled amplicon enrichment and bait hybridization. Sci Rep 13, 6461 (2023).
- Hourdel V, Kwasiborski A, Balière C, Matheus S, Batéjat CF, Manuguerra JC, Vanhomwegen J, Caro V. Rapid Genomic Characterization of SARS-CoV-2 by Direct Amplicon-Based Sequencing Through Comparison of MinION and Illumina iSeq100TM System. Front Microbiol. 2020
- Ke, S., Weiss, S.T. & Liu, YY. Dissecting the role of the human microbiome in COVID-19 via metagenome-assembled genomes. Nat Commun 13, 5235 (2022).


 Sample Submission Guidelines
Sample Submission Guidelines
