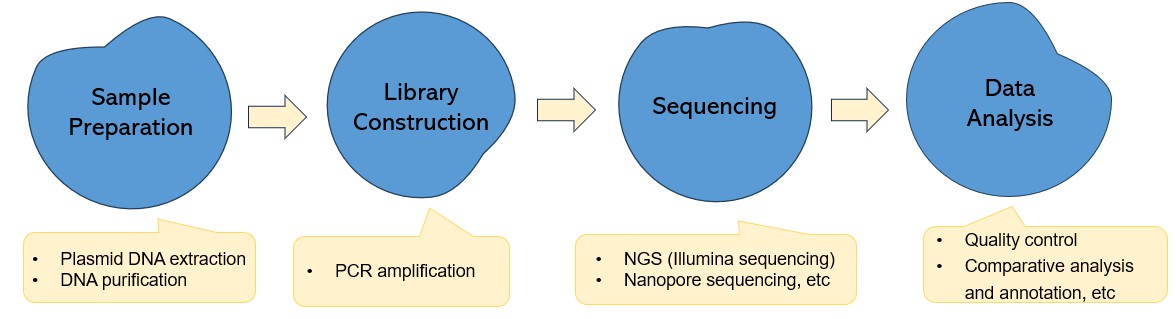
Plasmid Detection and Complete Plasmid DNA Sequencing
Introduction to Bacterial Plasmid
Bacterial plasmids are circular or linear double-stranded DNA molecules defined by their capability of autonomous replication in the hosts. They are critical sources for microbial evolution and genome innovation due to their ability to acquire foreign DNA sequences and transfer among bacteria and between distantly related organisms, like transferring from bacteria to eukaryotes via conjugation and mobilization. Bacterial plasmids undergo a higher frequency of genetic recombination than chromosomes. Plasmid genome, also known as the accessory or flexible genome, does not encode basic survival functions, but instead, it contributes to exploiting particular environmental niches, pathogenicity, degradation of aromatics, and antimicrobial resistance (AMR). Resistance to antibiotics has directly and sharply increased the number of multidrug-resistant bacteria. Thus, considerable effort has been made for plasmid detection and monitoring by using methods like complete plasmid DNA sequencing.
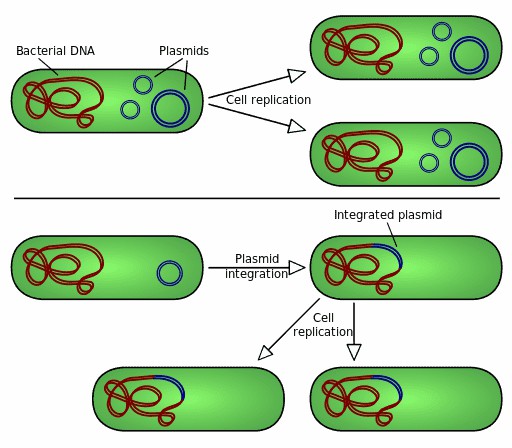 Figure 1. Autonomous replication and genomic integration of bacterial plasmids.
Figure 1. Autonomous replication and genomic integration of bacterial plasmids.
Services you may interested in
Plasmid Detection Methods
Plasmid detection stands as a fundamental technique widely employed in molecular biology research. Its primary objective is to ascertain the presence, size, and purity of plasmids, essential components in genetic engineering and molecular biology studies. Plasmid detection methodologies encompass a spectrum of approaches, including but not limited to PCR amplification, gel electrophoresis, fluorescence staining, optical mapping, and sequencing.
PCR-based replicon typing: This approach targets the conserved replicon sites of plasmid, and multiplex PCR can be expanded to simultaneously target many replicons. Although cheap and fast, this approach is hard to cover all novel plasmid groups.
Pulsed field gel electrophoresis (PFGE): This method separates digested DNA on a gel matrix by applying an electric field that periodically changes direction. However, it typically takes several days to reveal the size and number of plasmids in an isolate.
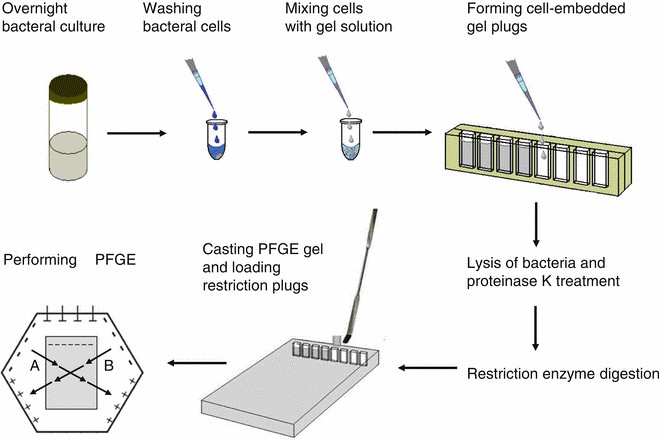 Figure 2. Pulsed field gel electrophoresis (Hu & Manos 2015)
Figure 2. Pulsed field gel electrophoresis (Hu & Manos 2015)
Optical mapping of plasmids: This method relying on the stretching of plasmid DNA can be used to depict the sequence of a plasmid. But optical mapping may not be suitable for the detection of short (< 50 Kb) plasmids.
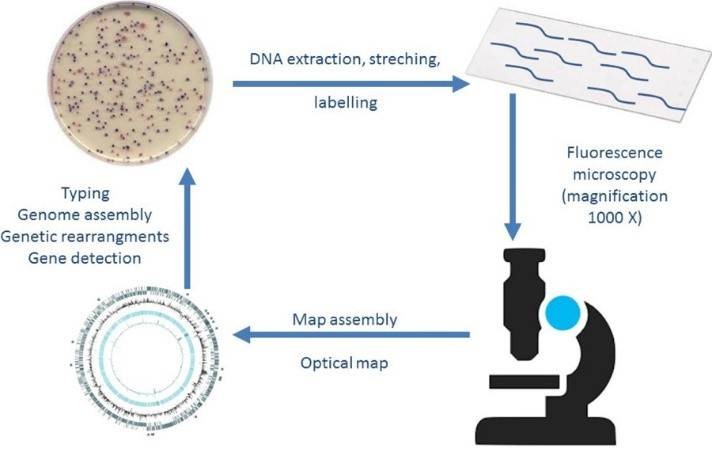 Figure 3. Optical mapping of plasmids (Bogas et al., 2017).
Figure 3. Optical mapping of plasmids (Bogas et al., 2017).
UV Spectrophotometry: UV spectrophotometry, a widely employed technique, serves to determine the concentration and purity of nucleic acid samples, including plasmid DNA. Nucleic acids absorb UV light around 260 nm. By measuring the absorbance at this wavelength, the concentration of DNA in the solution can be accurately determined.
Restriction Enzyme Digestion: Plasmid digestion with restriction enzymes is a pivotal step, wherein the expected enzyme cleavage pattern is employed to confirm the presence and size of plasmids.
Fluorescence Staining: Utilizing fluorescent dyes such as SYBR Green, fluorescence staining enables the visualization and quantification of plasmids by detecting their fluorescence signals.
Sequencing-based plasmid detection: With the development of high-throughput sequencing technologies, it is possible to obtain all the information contained in plasmids. Whole genome sequencing (WGS) can capture both bacterial and plasmid sequence data. Using algorithms such as PlasmidSeeker and plasmidSPAdes can extract and assemble plasmid data from WGS projects. However, these tools can only detect known plasmid sequence from raw WGS data. In order to comprehensively analyze plasmids, complete plasmid DNA sequencing provides a powerful tool for obtaining the full plasmid sequence.
What Is Complete Plasmid DNA Sequencing?
Complete Plasmid DNA Sequencing, also known as Whole Plasmid Sequencing or Full Plasmid Sequencing, refers to the process of determining and analysing the entire DNA sequence of a plasmid. This sequencing method enables a comprehensive overview of the complete configuration of the plasmid, incorporating genes, primers, promoters, and other critical sequences it may contain. Leveraging the capabilities of Next Generation Sequencing (NGS), PacBio sequencing, or Nanopore sequencing, it permits the sequencing of all nucleotide sequences within any given plasmid. This approach facilitates a comprehensive characterization of unknown plasmids and thorough verification of known ones. Notably, this method does not require primers or rely on a reference database.
The role of Complete Plasmid DNA Sequencing:
Verification of Sequence Integrity: During the process of cloning or amplification, plasmids may undergo genetic modifications, potentially leading to unwanted sequence errors or mutations.
Quality Control in Cloning and Engineering: Sequencing plasmids serves as a quality control step, substantiating the accuracy of cloned inserts, confirming the absence of unwanted mutations, and ensuring the integrity of the experimental system.
Optimization of Experimental Design: Accurate knowledge of a plasmid's sequence enables researchers to design experiments more efficiently. The full comprehension of a plasmid's composition provides fundamental insights that could guide designed manipulations and predict their possible outcomes.
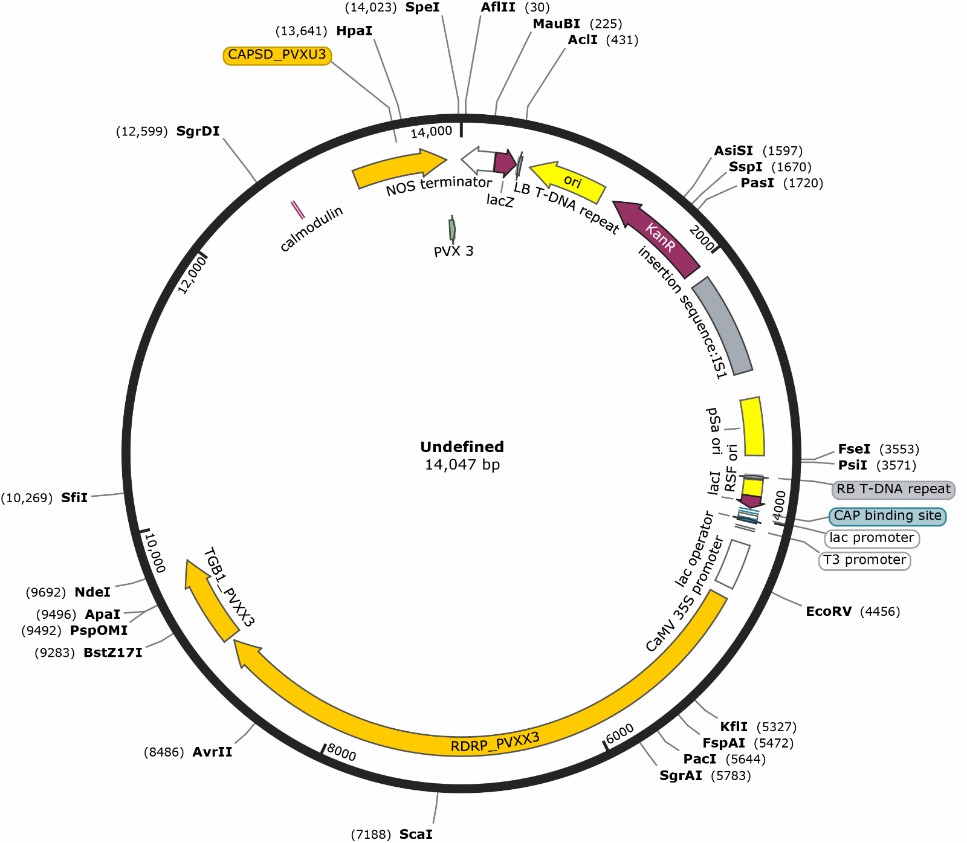 Figure 4. Plasmid circle diagram
Figure 4. Plasmid circle diagram
Process of Complete Plasmid DNA Sequencing
The workflow for complete plasmid sequencing includes plasmid DNA preparation and quality control (QC), library construction and QC, sequencing, and bioinformatics analysis (including data QC, de novo assembly and further analysis). For NGS, it is difficult to achieve contiguous assemblies of plasmids as the read length, typically less than ∼400 to 500 bp, cannot cover the full-length of most mobile genetic elements. Thus, assemblies of plasmids from NGS data are often fragmented or incomplete. Third-generation sequencing, also known as long-read sequencing, includes PacBio and Nanopore sequencing, delivering rapid plasmid sequencing and allowing full assembly of plasmids to closure in many cases. The primary advantages of employing Next-Generation Sequencing (NGS) for complete plasmid sequencing lie in its superior precision and high throughput capabilities. Conversely, the primary benefits of Nanopore technology are manifested in its optimal performance with high GC content and Inverted Terminal Repeat (ITR) structures. Additional attributes include extensive read lengths and shortened turnaround times.
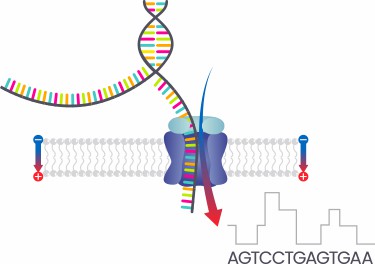
Several precautions should be taken during Complete Plasmid DNA Sequencing:
- Employ suitable extraction methods and Plasmid DNA extraction kits to avoid contamination and degradation. Subsequently, an accurate measurement of the plasmid DNA concentration should be carried out using colorimetric or fluorescent methods, ensuring the use of identical DNA quantities in subsequent experiments.
- Based on the experimental requirements and the type of samples, the appropriate library construction method, such as PCR amplification or transposon method, should be selected. When using PCR amplification, the PCR conditions should be optimized to prevent amplification biases and the introduction of hybrid products, thus ensuring a uniform length of plasmid DNA fragments in the library.
- Quality control software should be applied to filter and trim sequencing data, eliminating low-quality reads and adapter sequences to enhance the accuracy of subsequent analysis.
- Suitable control groups, including positive, negative, and blank controls, should be established. This ensures the reliability and preciseness of the experimental results. Repetition of experiments strengthens validation; thereby enhancing the credibility of the results.
Application Of Complete Plasmid DNA Sequencing
In genetic engineering and molecular biology research, complete plasmid sequencing is employed to verify the accuracy and integrity of plasmid constructions. Through sequencing analysis, one can affirm the presence of desired genetic sequences, promoters, tags, etc., within the plasmid, thereby ensuring conformance with design anticipations. As pertains to gene function research, complete plasmid sequencing can illuminate the functionalities and regulatory mechanisms of genes within the plasmid. By analysing plasmid sequences, we can identify the genes carried by the plasmid and further our understanding of these genes' functions, expressions patterns, and regulatory networks within the cell.
In disease research, complete plasmid sequencing is utilised to identify plasmid variations and mutations associated with disease. By comparing plasmid sequences from patient samples with control samples, potential gene markers connected to the onset and progression of disease can be identified. With regard to drug development and targeted therapy, complete plasmid sequencing can aid in the identification of potential drug targets. By analysing the genes and regulatory elements within the plasmid, novel targets associated with specific diseases can be identified, offering new directions for drug design and development.
In the field of environmental microbiology research, complete plasmid sequencing serves as a valuable tool for analyzing the composition and functionality of microbial plasmids within environmental samples. By scrutinizing the microbial plasmid repertoire and its functional attributes, this approach contributes to a comprehensive understanding of microbial metabolic traits, ecological roles, and adaptive strategies within diverse environmental contexts. Furthermore, in the realm of genetic studies, complete plasmid sequencing facilitates the elucidation of plasmid genetic diversity and evolutionary relationships across different species and populations. This aids in unraveling the roles of plasmids in genetic transmission and species adaptation processes, thus enriching our insights into evolutionary dynamics and genetic mechanisms underlying microbial communities.
Complete Plasmid Sequencing and Sanger sequencing
Sanger sequencing and complete plasmid sequencing represent two widely implemented techniques for DNA sequencing, each offering unique characteristics while still bearing an integral relationship with one another. Sanger sequencing functions primarily on the chain termination method, tailored particularly for sequencing shorter DNA strands such as specific genes or primer sequences. This technique also finds utility in the detection of specific gene mutations. In terms of cost-effectiveness, Sanger sequencing provides a relatively affordable solution, making it suitable for smaller scale sequencing projects.
Complete plasmid sequencing represents a sophisticated technique to sequence the entire DNA of a plasmid. This technique primarily harnesses the power of next-generation sequencing (NGS) or long-read sequencing to ascertain the sequence of the complete plasmid DNA. Conventionally, the sequencing method encompasses amplification of plasmid DNA through polymerase chain reaction (PCR), followed by high-throughput sequencing of the amplified products enabling the retrieval of the full plasmid sequence. This robust method is aptly suited for a diverse range of applications that include plasmid construction and validation, disease research, and environmental microbiology studies. Notwithstanding the relatively high cost associated with complete plasmid sequencing, this technique facilitates access to the entire sequence of a plasmid, making it an invaluable tool for expansive sequencing projects requiring comprehensive analyses.
While both Sanger sequencing and complete plasmid sequencing cater to the determination of DNA sequences and can obtain DNA sequence information via sequencing machinery, they can effectively complement each other under certain conditions. For instance, during the process of plasmid construction, Sanger sequencing can be employed to verify specific regions of the plasmid, whereas complete plasmid sequencing can ascertain the entirety and precision of the plasmid.
References:
- Roosaare M, Puustusmaa M, Möls M, et al. PlasmidSeeker: identification of known plasmids from bacterial whole genome sequencing reads. PeerJ, 2018, 6: e4588.
- Gutiérrez-Barranquero J A, Cazorla F M, de Vicente A, et al. Complete sequence and comparative genomic analysis of eight native Pseudomonas syringae plasmids belonging to the pPT23A family. BMC genomics, 2017, 18(1): 365.
- Jackson R W, Vinatzer B, Arnold D L, et al. The influence of the accessory genome on bacterial pathogen evolution. Mobile genetic elements, 2011, 1(1): 55-65.
- Bogas D, Nyberg L, Pacheco R, et al. Applications of optical DNA mapping in microbiology. BioTechniques, 2017, 62(6): 255-267.
- Lemon J K, Khil P P, Frank K M, et al. Rapid nanopore sequencing of plasmids and resistance gene detection in clinical isolates. Journal of clinical microbiology, 2017, 55(12): 3530-3543.
- Hu H, Manos J. Pulsed-field gel electrophoresis of Pseudomonas aeruginosa//Pulse Field Gel Electrophoresis. Humana Press, New York, NY, 2015: 157-170.


 Sample Submission Guidelines
Sample Submission Guidelines
