Ribosome Profiling: Definition, Applications, Principles, and Workflow
What Is Ribosome Profiling?
Ribosome profiling, also known as Ribo-seq, is a recently developed high-throughput sequencing technique based on the sequencing of ribosome-protected mRNA fragments. Ribosome profiling allows the detailed and accurate measurement of cellular translation, even in vivo. Ribo-seq enables the monitoring of cellular translation processes, determination for actively translated proteins in a cell, and prediction of protein abundance, thus helping define the proteome of complex organisms. Furthermore, Ribo-seq can provide insights into the mechanism of translational regulation. For example, it can be used to detect regulatory translational pauses and translated upstream ORFs, and monitor translation processes mediated by subsets of ribosomes. Ribosome profiling is the first method available for the annotation of translated open reading frames (ORFs) and has enabled the discovery of many novel or alternative protein products.
Applications of ribosome profiling:
- Identify translated sequences within the complicated transcriptome
- Map sites of translation initiation
- Measure differential gene expression at the level of mRNA translation
- Identify novel protein coding genes and ribosome pausing
- Provide data for quantitative models of translation
- Provide insights into the mechanism of protein synthesis, translation control, and process of viral infection
Principles of Ribosome Profiling
Ribosome profiling takes the advantage of the fact that ribosomes engaging in translation can protect about 30 nucleotides of an mRNA fragment against RNAse digestion. This technique first exploits the classical molecular method of ribosome footprinting. In this method, in vitro translated mRNAs are treated with nuclease to destroy unprotected regions. The mRNAs protected by the ribosome can be mapped back to the original mRNA to determine the exact location of the translating ribosome. Ribosome profiling extends the method of ribosome footprinting by mapping and measuring the full complement of ribosome footprints to quantify novel protein synthesis and annotate coding regions globally. Advances in the sequencing technology enable deep and accurate analysis of all translating ribosomes by mRNA-seq.
Workflow of Ribosome Profiling
Next-generation sequencing (NGS)-based ribosome profiling requires preparation of a physiological sample, in vivo capture of translating ribosomes and mRNAs, lysis, nuclease digestion (typically RNase I or micrococcal nuclease) to obtain ribosome-protected fragments, isolation of ribosomes and subsequently, ribosome footprints, which are converted to a strand-specific library and subjected to NGS. Sequences from the fragments are then mapped against the appropriate reference genome. Ribosome profiling provides genome-wide information on protein synthesis (GWIPS) in vivo.
By comparing the rates of protein synthesis and the abundance of mRNAs, the translational efficiency for each mRNA can be determined.
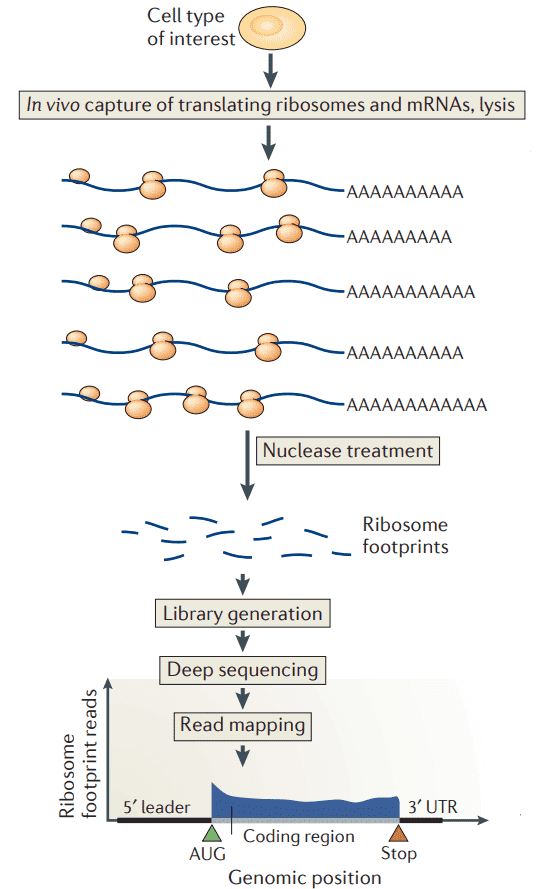 Figure 1. Workflow for ribosome profiling (Brar and Weissman 2015).
Figure 1. Workflow for ribosome profiling (Brar and Weissman 2015).
References:
- Brar G A, Weissman J S. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nature reviews Molecular cell biology, 2015, 16(11): 651-664.
- Ingolia N T. Ribosome profiling: new views of translation, from single codons to genome scale. Nature Reviews Genetics, 2014, 15(3): 205-213.


 Sample Submission Guidelines
Sample Submission Guidelines
