Million-Year-Old DNA Provides a Glimpse of Mammoth Evolution
February 17, 2021
In 2013, DNA from a horse that lived sometime between 560,000 and 780,000 years ago was sequenced. It was the most ancient DNA sample ever analysed. But that record has just been smashed by van der Valk and colleagues, writing in Nature. The authors retrieved DNA from the molars of three mammoths found in northeast Siberia, two of which lived more than one million years (Myr) ago.
The authors isolated the DNA from molars that had been previously collected from the Siberian permafrost. They relied on methods that maximize the recovery of short fragments of residual DNA. The cold temperatures had lessened degradation of the DNA across geological timescales.
The authors dated the mammoths using biostratigraphy, in which faunal remains at the sites where the molars were collected are correlated with fauna at sites for which absolute dates are available. They also estimated the antiquity of the specimens by molecular dating of DNA in a cellular organelle called the mitochondrion, because a higher percentage of the mitochondrial genome was covered by sequencing than was that of nuclear DNA (although similar date estimates were obtained using nuclear DNA from the two more recent specimens).
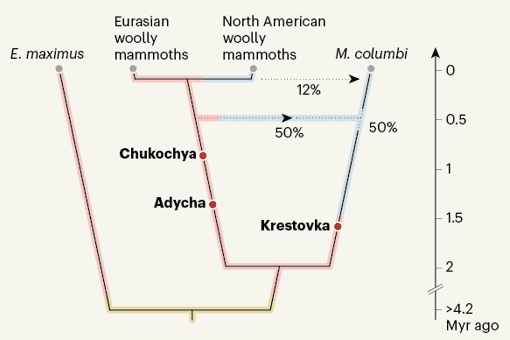
The mitogenome data revealed that the most recent of the three mammoth specimens, dubbed Chukochya, lived more than 680,000 years ago (for comparison, the iconic woolly mammoth, Mammuthus primigenius, first appeared in the fossil record around 700,000 years ago). In a phylogenetic tree made using nuclear DNA, Chukochya fell outside a group comprising all woolly mammoths from the Late Pleistocene (129,000–12,000 years ago). This finding is consistent with the morphology of the Chukochya molar, which identified it as an early form of woolly mammoth.
The second-oldest mammoth, called Adycha, lived about 1.34 Myr ago during the Early Pleistocene (which spanned from about 2.58 million to 773,000 years ago). The authors found that Adycha belonged to a population ancestral to woolly mammoths, and which lived before Chukochya. There were substantial differences between the molar of Adycha and those of Chukochya and more-recent woolly mammoths, in terms of enamel thickness, number and density of enamel plates, and height of the crowns. We do not yet have a good enough understanding of the developmental genetic programs that underlie these and other morphological traits to be able to identify the genomic changes responsible. Going forward, a better understanding of the genetics of skull and tooth development might enable a closer braiding of mammoth genomics with palaeontology.
The authors went on to compare the genomes of these ancient specimens with those of the animals' woolly mammoth descendants, to examine how these mammoths had adapted to their cold Siberian environment. Many genetic variants thought to be the result of adaptation to northern latitudes have been identified in woolly mammoths by comparing their genomes with those of African savannah (Loxodonta africana) and Asian (Elephas maximus) elephants, members of the same mammalian family. Of these variants, van der Valk et al. showed that 87% were already present in Adycha and 89% in Chukochya. This is not surprising, because any lineage preserved in permafrost must already have been adapted to frigid climates. However, the authors also found evidence for further adaptation as the mammoth lineage evolved. For example, the gene TRPV3, involved in sensing temperature, carried more variants in Late Pleistocene woolly mammoths than in the ancestral Chukochya.
The most ancient mammoth was Krestovka, estimated by mitogenome dating to have lived about 1.65 Myr ago (although biostratigraphy suggested a slightly more recent date). A phylogenetic tree indicated that Krestovka was not from a population ancestral to woolly mammoths. Instead, it was part of a lineage that split from the Adycha–Chukochya–woolly mammoth line roughly 2 Myr ago. The researchers propose that Krestovka was ancestral to the mammoths that entered North America about 1.5 Myr ago and gave rise to the Columbian mammoth (Mammuthus columbi) in regions of mild climate in North and Central America.
The authors also found, in the nuclear genomes of Columbian mammoths, the signatures of two admixture (interbreeding) events between the Krestovka and woolly mammoth lineages. The second of these seems to have occurred after woolly mammoths entered North America about 100,000 years ago. In this event, about 12% of the Columbian mammoth genome was replaced by DNA from Late Pleistocene woolly mammoths.
The remainder of the Columbian mammoth genome showed equal contributions from the Krestovka and woolly mammoth lineages, indicative of an earlier admixture event. A 50:50 split of this type might be considered surprising, given that the contributions of two ancestral populations could be in any proportions — 80:30, for instance, or 10:90. The split could be coincidence, but van der Valk and colleagues suggest the possibility of hybrid speciation, in which the offspring of crosses between the two lineages do not themselves breed with either parental lineage.
The authors propose that the initial hybridization occurred around 420,000 years ago, because at this time there seems to have been a transfer of mitogenomes from a woolly mammoth lineage to the Columbian mammoth. However, the ancestors of Columbian mammoths entered North America long before this date, and woolly mammoths long after, making it difficult to harmonize the date with the fossil record. Mitogenomes transfer readily between species in the elephant family, so perhaps the phylogeny of Late Pleistocene mammoths reflects more events than just a single interspecies transfer. Alternatively, the hybridization evident in the nuclear genome might not have occurred at the time estimated using mitogenomes. If DNA could be obtained from Early or Middle Pleistocene mammoth specimens in the permafrost of North America, this might shed further light on the origins of Columbian mammoths.
Many present-day animal species arose during or after the Early Pleistocene. The ability to retrieve DNA from Early Pleistocene specimens means that genomic changes in some lineages can now be tracked across deep time, providing insights into the evolution of modern species. Modern genomes are often used to infer the demographic history of populations across hundreds of thousands of years; these inferences should now be tested for accuracy by examining specimens across real time.
Tracking mitochondrial and nuclear genetic changes across deep time could also reveal the role (if any) of mitochondrial–nuclear interactions in mammoth evolution. When mitochondria are switched between cells from different species in vitro, interactions between the proteins encoded by mitochondrial and nuclear genes can be disrupted. It is conceivable that such disruption could drive the evolution or disappearance of mitogenome lineages. This avenue of investigation is especially pertinent, given that van der Valk and colleagues relied on the transfer of mitogenomes to date the hybridization that gave rise to Columbian mammoths.
Finally, biostratigraphy focuses on the distribution and morphology of small species such as lemmings, pikas and voles. It should now be possible to add a DNA component to biostratigraphy, through genomic analyses of small mammals across sites. Genomics has been pushed into deep time by the giants of the Ice Age — the wee mammals that surrounded them might soon also have their day.
More info at: https://www.nature.com/articles/d41586-021-00348-w


 Sample Submission Guidelines
Sample Submission Guidelines
